Predominance of six different hexanucleotide recoding signals 3' of read-through stop codons
- PMID: 11972340
- PMCID: PMC113845
- DOI: 10.1093/nar/30.9.2011
Predominance of six different hexanucleotide recoding signals 3' of read-through stop codons
Abstract
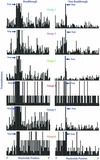
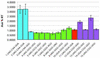
Redefinition of UAG, UAA and UGA to specify a standard amino acid occurs in response to recoding signals present in a minority of mRNAs. This 'read-through' is in competition with termination and is utilized for gene expression. One of the recoding signals known to stimulate read-through is a hexanucleotide sequence of the form CARYYA 3' adjacent to the stop codon. The present work finds that of the 91 unique viral sequences annotated as read-through, 90% had one of six of the 64 possible codons immediately 3' of the read-through stop codon. The relative efficiency of these read-through contexts in mammalian tissue culture cells has been determined using a dual luciferase fusion reporter. The relative importance of the identity of several individual nucleotides in the different hexanucleotides is complex.
Figures
References
-
- Low S.C. and Berry,M.J. (1996) Knowing when not to stop: selenocysteine incorporation in eukaryotes. Trends Biochem. Sci., 21, 203–208. - PubMed
-
- Robinson D.N. and Cooley,L. (1997) Examination of the function of two kelch proteins generated by stop codon suppression. Development, 124, 1405–1417. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

