Genomic repeats, genome plasticity and the dynamics of Mycoplasma evolution
- PMID: 11972343
- PMCID: PMC113839
- DOI: 10.1093/nar/30.9.2031
Genomic repeats, genome plasticity and the dynamics of Mycoplasma evolution
Abstract
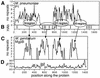
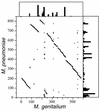
Mycoplasmas evolved by a drastic reduction in genome size, but their genomes contain numerous repeated sequences with important roles in their evolution. We have established a bioinformatic strategy to detect the major recombination hot-spots in the genomes of Mycoplasma pneumoniae, Mycoplasma genitalium, Ureaplasma urealyticum and Mycoplasma pulmonis. This allowed the identification of large numbers of potentially variable regions, as well as a comparison of the relative recombination potentials of different genomic regions. Different trends are perceptible among mycoplasmas, probably due to different functional and structural constraints. The largest potential for illegitimate recombination in M.pulmonis is found at the vsa locus and its comparison in two different strains reveals numerous changes since divergence. On the other hand, the main M.pneumoniae and M.genitalium adhesins rely on large distant repeats and, hence, homologous recombination for variation. However, the relation between the existence of repeats and antigenic variation is not necessarily straightforward, since repeats of P1 adhesin were found to be anti-correlated with epitopes recognized by patient antibodies. These different strategies have important consequences for the structures of genomes, since large distant repeats correlate well with the major chromosomal rearrangements. Probably to avoid such events, mycoplasmas strongly avoid inverse repeats, in comparison to co-oriented repeats.
Figures


References
-
- Krause D.C. and Balish,M.F. (2001) Structure, function and assembly of the terminal organelle of Mycoplasma pneumoniae. FEMS Microbiol. Lett., 198, 1–7. - PubMed
-
- Kenri T., Taniguchi,R., Sasaki,Y., Okazaki,N., Narita,M., Izumikawa,K., Umetsu,M. and Sasaki,T. (1999) Identification of a new variable sequence in the P1 cytadhesin gene of Mycoplasma pneumoniae: evidence for the generation of antigenic variation by DNA recombination between repetitive sequences. Infect. Immun., 67, 4557–4562. - PMC - PubMed
-
- Fraser C.M., Gocayne,J.D., White,O., Adams,M.D., Clayton,R.A., Fleischmann,R.D., Bult,C.J., Kerlavage,A.R., Sutton,G., Kelley,J.M. et al. (1995) The minimal gene complement of Mycoplasma genitalium. Science, 270, 397–403. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

