Partitioning of bacterial communities between seawater and healthy, black band diseased, and dead coral surfaces
- PMID: 11976091
- PMCID: PMC127591
- DOI: 10.1128/AEM.68.5.2214-2228.2002
Partitioning of bacterial communities between seawater and healthy, black band diseased, and dead coral surfaces
Abstract
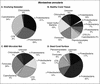
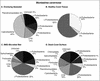
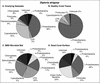
Distinct partitioning has been observed in the composition and diversity of bacterial communities inhabiting the surface and overlying seawater of three coral species infected with black band disease (BBD) on the southern Caribbean island of Curaçao, Netherlands Antilles. PCR amplification and sequencing of bacterial 16S rRNA genes (rDNA) with universally conserved primers have identified over 524 unique bacterial sequences affiliated with 12 bacterial divisions. The molecular sequences exhibited less than 5% similarity in bacterial community composition between seawater and the healthy, black band diseased, and dead coral surfaces. The BBD bacterial mat rapidly migrates across and kills the coral tissue. Clone libraries constructed from the BBD mat were comprised of eight bacterial divisions and 13% unknowns. Several sequences representing bacteria previously found in other marine and terrestrial organisms (including humans) were isolated from the infected coral surfaces, including Clostridium spp., Arcobacter spp., Campylobacter spp., Cytophaga fermentans, Cytophaga columnaris, and Trichodesmium tenue.
Figures

References
-
- Acinas, S. G., F. Rodriguez-Valera, and C. Pedros-Alio. 1997. Spatial and temporal variation in marine bacterioplankton diversity as shown by RFLP fingerprinting of PCR amplified 16S rRNA. FEMS Microb. Ecol. 24:27-40.
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Meyers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 59:143-169. - PubMed
-
- Anderson, J. I. W., and D. A. Conroy. 1969. The pathogenic myxobacteria with special reference to fish diseases. J. Appl. Bacteriol. 32:30-39. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

