Bacillus probiotics: spore germination in the gastrointestinal tract
- PMID: 11976107
- PMCID: PMC127533
- DOI: 10.1128/AEM.68.5.2344-2352.2002
Bacillus probiotics: spore germination in the gastrointestinal tract
Abstract
Spores of Bacillus species are being used commercially as probiotics and competitive exclusion agents. Unlike the more commonly used Lactobacillus-type probiotics, spores are dormant life forms. To address how spore probiotics might function we have investigated whether spores can germinate in the gastrointestinal tract by using a murine model. Using a genetically engineered chimeric gene, ftsH-lacZ, which is strongly expressed only in vegetative cells, we have developed a sensitive competitive reverse transcription-PCR assay which has enabled detection of as few as 10(2) vegetative bacteria in the mouse gut. Using this method we have administered doses of ftsH-lacZ spores to groups of mice and shown that spores can germinate in significant numbers in the jejunum and ileum. The levels of detection we obtained suggest that spores may colonize the small intestine, albeit briefly.
Figures

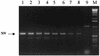
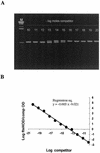




References
-
- Atlas, R. M. 1999. Probiotics: snake oil for the new millenium? Environ. Microbiol. 1:375-382. - PubMed
-
- Caruso, A., G. Flamminio, S. Folghera, L. Peroni, I. Foresti, A. Balsari, and A. Turano. 1993. Expression of activation markers on peripheral-blood lymphocytes following oral administration of Bacillus subtilis spores. Int. J. Immunopharmacol. 15:87-92. - PubMed
-
- Chomczynski, P., and N. Sacchi. 1987. Single step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. - PubMed
-
- Ciprandi, G., A. Scordamaglia, D. Venuti, M. Caria, and G. W. Canonica. 1986. In vitro effects of Bacillus subtilis on the immune response. Chemioterapia 5:404-407. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

