Cre-loxP recombination system for large genome rearrangements in Lactococcus lactis
- PMID: 11976109
- PMCID: PMC127585
- DOI: 10.1128/AEM.68.5.2359-2367.2002
Cre-loxP recombination system for large genome rearrangements in Lactococcus lactis
Abstract
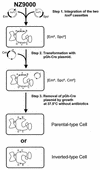
We have used a new genetic strategy based on the Cre-loxP recombination system to generate large chromosomal rearrangements in Lactococcus lactis. Two loxP sites were sequentially integrated in inverse order into the chromosome either at random locations by transposition or at fixed points by homologous recombination. The recombination between the two chromosomal loxP sites was highly efficient (approximately 1 x 10(-1)/cell) when the Cre recombinase was provided in trans, and parental- or inverted-type chromosomal structures were isolated after removal of the Cre recombinase. The usefulness of this approach was demonstrated by creating three large inversions of 500, 1,115, and 1,160 kb in size that modified the lactococcal genome organization to different extents. The Cre-loxP recombination system described can potentially be used for other gram-positive bacteria without further modification.
Figures




References
-
- Abremski, K., R. Hoess, and N. Sternberg. 1983. Studies on the properties of P1 site-specific recombination: evidence for topologically unlinked products following recombination. Cell 32:1301-1311. - PubMed
-
- Altier, C., and M. Suyemoto. 1999. A recombinase-based selection of differentially expressed bacterial genes. Gene 240:99-106. - PubMed
-
- Aranda, M., C. Kanellopoulou, N. Christ, M. Peitz, K. Rajewsky, and P. Droge. 2001. Altered directionality in the Cre-loxP site-specific recombination pathway. J. Mol. Biol. 311:453-459. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

