Assessing the role of Pseudomonas aeruginosa surface-active gene expression in hexadecane biodegradation in sand
- PMID: 11976128
- PMCID: PMC127520
- DOI: 10.1128/AEM.68.5.2509-2518.2002
Assessing the role of Pseudomonas aeruginosa surface-active gene expression in hexadecane biodegradation in sand
Abstract
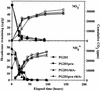
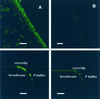
Low pollutant substrate bioavailability limits hydrocarbon biodegradation in soils. Bacterially produced surface-active compounds, such as rhamnolipid biosurfactant and the PA bioemulsifying protein produced by Pseudomonas aeruginosa, can improve bioavailability and biodegradation in liquid culture, but their production and roles in soils are unknown. In this study, we asked if the genes for surface-active compounds are expressed in unsaturated porous media contaminated with hexadecane. Furthermore, if expression does occur, is biodegradation enhanced? To detect expression of genes for surface-active compounds, we fused the gfp reporter gene either to the promoter region of pra, which encodes for the emulsifying PA protein, or to the promoter of the transcriptional activator rhlR. We assessed green fluorescent protein (GFP) production conferred by these gene fusions in P. aeruginosa PG201. GFP was produced in sand culture, indicating that the rhlR and pra genes are both transcribed in unsaturated porous media. Confocal laser scanning microscopy of liquid drops revealed that gfp expression was localized at the hexadecane-water interface. Wild-type PG201 and its mutants that are deficient in either PA protein, rhamnolipid synthesis, or both were studied to determine if the genetic potential to make surface-active compounds confers an advantage to P. aeruginosa biodegrading hexadecane in sand. Hexadecane depletion rates and carbon utilization efficiency in sand culture were the same for wild-type and mutant strains, i.e., whether PG201 was proficient or deficient in surfactant or emulsifier production. Environmental scanning electron microscopy revealed that colonization of sand grains was sparse, with cells in small monolayer clusters instead of multilayered biofilms. Our findings suggest that P. aeruginosa likely produces surface-active compounds in sand culture. However, the ability to produce surface-active compounds did not enhance biodegradation in sand culture because well-distributed cells and well-distributed hexadecane favored direct contact to hexadecane for most cells. In contrast, surface-active compounds enable bacteria in liquid culture to adhere to the hexadecane-water interface when they otherwise would not, and thus production of surface-active compounds is an advantage for hexadecane biodegradation in well-dispersed liquid systems.
Figures



Similar articles
-
Rhamnolipid biosurfactant enhancement of hexadecane biodegradation by Pseudomonas aeruginosa.Mol Mar Biol Biotechnol. 1995 Dec;4(4):331-7. Mol Mar Biol Biotechnol. 1995. PMID: 8541984
-
Rhamnolipid (biosurfactant) effects on cell aggregation and biodegradation of residual hexadecane under saturated flow conditions.Appl Environ Microbiol. 1997 Sep;63(9):3622-7. doi: 10.1128/aem.63.9.3622-3627.1997. Appl Environ Microbiol. 1997. PMID: 9293014 Free PMC article.
-
Isolation and characterization of a regulatory gene affecting rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa.J Bacteriol. 1994 Apr;176(7):2044-54. doi: 10.1128/jb.176.7.2044-2054.1994. J Bacteriol. 1994. PMID: 8144472 Free PMC article.
-
Role of rhamnolipid biosurfactants in the uptake and mineralization of hexadecane in Pseudomonas aeruginosa.J Appl Microbiol. 2000 Jul;89(1):158-68. doi: 10.1046/j.1365-2672.2000.01104.x. J Appl Microbiol. 2000. PMID: 10945793
-
Quorum sensing: implications on rhamnolipid biosurfactant production.Biotechnol Genet Eng Rev. 2010;27:159-84. doi: 10.1080/02648725.2010.10648149. Biotechnol Genet Eng Rev. 2010. PMID: 21415897 Review.
Cited by
-
Separation of Bacteria, Protozoa and Carbon Nanotubes by Density Gradient Centrifugation.Nanomaterials (Basel). 2016;6(10):181. doi: 10.3390/nano6100181. Epub 2016 Oct 12. Nanomaterials (Basel). 2016. PMID: 27917301 Free PMC article.
-
Recent advances in petroleum microbiology.Microbiol Mol Biol Rev. 2003 Dec;67(4):503-49. doi: 10.1128/MMBR.67.4.503-549.2003. Microbiol Mol Biol Rev. 2003. PMID: 14665675 Free PMC article. Review.
-
Microbial community composition and denitrifying enzyme activities in salt marsh sediments.Appl Environ Microbiol. 2008 Dec;74(24):7585-95. doi: 10.1128/AEM.01221-08. Epub 2008 Oct 31. Appl Environ Microbiol. 2008. PMID: 18978080 Free PMC article.
-
Soil bioremediation approaches for petroleum hydrocarbon polluted environments.AIMS Microbiol. 2017 Jan 19;3(1):25-49. doi: 10.3934/microbiol.2017.1.25. eCollection 2017. AIMS Microbiol. 2017. PMID: 31294147 Free PMC article. Review.
-
Dispersion of TiO₂ nanoparticle agglomerates by Pseudomonas aeruginosa.Appl Environ Microbiol. 2010 Nov;76(21):7292-8. doi: 10.1128/AEM.00324-10. Epub 2010 Sep 17. Appl Environ Microbiol. 2010. PMID: 20851981 Free PMC article.
References
-
- Abu-Ruwaida, A. S., I. M. Banat, S. Haditirto, A. Salem, and M. Kadri. 1991. Isolation of biosurfactant-producing bacteria, product characterization, and evaluation. Acta Biotechnol. 11:315-324.
-
- Akit, J., D. G. Cooper, K. I. Manninen, and J. E. Zajic. 1981. Investigation of potential biosurfactant production among phytopathogenic Corynebacteria and related soil microbes. Curr. Microbiol. 6:145-150.
-
- Alexander, M. 1999. Biodegradation and bioremediation, 2nd ed. Academic Press, San Diego, Calif.
-
- Bai, G., M. L. Brusseau, and R. M. Miller. 1997. Biosurfactant-enhanced removal of residual hydrocarbon from soil. J. Contam. Hydrol. 25:157-170.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

