Genomic sequence and evolution of marine cyanophage P60: a new insight on lytic and lysogenic phages
- PMID: 11976141
- PMCID: PMC127578
- DOI: 10.1128/AEM.68.5.2589-2594.2002
Genomic sequence and evolution of marine cyanophage P60: a new insight on lytic and lysogenic phages
Abstract
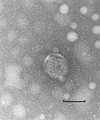
The genome of cyanophage P60, a lytic virus which infects marine Synechococcus WH7803, was completely sequenced. The P60 genome contained 47,872 bp with 80 potential open reading frames that were mostly similar to the genes found in lytic phages like T7, phi-YeO3-12, and SIO1. The DNA replication system, consisting of primase-helicase and DNA polymerase, appeared to be more conserved in podoviruses than in siphoviruses and myoviruses, suggesting that DNA replication genes could be the critical elements for lytic phages. Strikingly high sequence similarities in the regions coding for nucleotide metabolism were found between cyanophage P60 and marine unicellular cyanobacteria.
Figures


References
-
- Bergh, O., K. Y. Borsheim, G. Bratbak, and M. Heldal. 1989. High abundance of viruses found in aquatic environments. Nature 340:467-468. - PubMed
-
- Brussow, H., and F. Desiere. 2001. Comparative phage genomics and the evolution of Siphoviridae: insights from dairy phages. Mol. Microbiol. 39:213-222. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

