Structural basis of VDR-DNA interactions on direct repeat response elements
- PMID: 11980721
- PMCID: PMC125986
- DOI: 10.1093/emboj/21.9.2242
Structural basis of VDR-DNA interactions on direct repeat response elements
Abstract
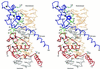
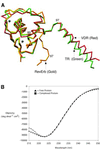
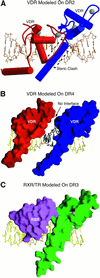
The vitamin D receptor (VDR) forms homo- or heterodimers on response elements composed of two hexameric half-sites separated by 3 bp of spacer DNA. We describe here the crystal structures at 2.7-2.8 A resolution of the VDR DNA-binding region (DBD) in complex with response elements from three different promoters: osteopontin (SPP), canonical DR3 and osteocalcin (OC). These structures reveal the chemical basis for the increased affinity of VDR for the SPP response element, and for the poor stability of the VDR-OC complex, relative to the canonical DR3 response element. The homodimeric protein-protein interface is stabilized by van der Waals interactions and is predominantly non-polar. An extensive alpha-helix at the C-terminal end of the VDR DBD resembles that found in the thyroid hormone receptor (TR), and suggests a mechanism by which VDR and TR discriminate among response elements. Selective structure-based mutations in the asymmetric homodimeric interface result in a VDR DBD protein that is defective in homodimerization but now forms heterodimers with the 9-cis retinoic acid receptor (RXR) DBD.
Figures


References
-
- Bouillon R., Okamura,W.H. and Norman,A.W. (1995) Structure–function relationships in the vitamin D endocrine system. Endocr. Rev., 16, 200–257. - PubMed
-
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr., D54, 905–921. - PubMed
-
- Carson M. (1991) Ribbons 2.0. J. Appl. Crystallogr., 24, 958–961.
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

