Molecular dynamics simulations of protein folding from the transition state
- PMID: 11983864
- PMCID: PMC124469
- DOI: 10.1073/pnas.092686399
Molecular dynamics simulations of protein folding from the transition state
Abstract
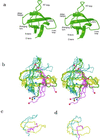
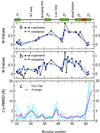
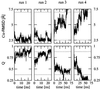
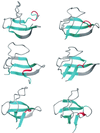
Putative transition-state ensemble (TSE) conformations of src SH3 were identified by monitoring the deviation from the experimental phi values along molecular dynamics (MD) simulations of unfolding. Sixty MD trajectories (for a total of about 7 micros) were then started from the putative TSE. About one-half of the 60 runs reached the folded state while unfolding was observed in the remaining half of the runs. This result validates phi-value analysis as an approach to obtain structural information on the transition state. It also demonstrates that an atomic resolution description of the TSE can be extracted from MD simulations. All conformations in the TSE have the central three-stranded beta-sheet formed in agreement with experimental data. An elongation of strand beta 2 as well as non-native side-chain interactions between the diverging turn and the distal hairpin are observed. The simulation results indicate that the tight packing of the side chains between the diverging turn and the distal hairpin is a necessary condition for rapid folding. Contacts between residues in the most structured element of the TSE, the central beta-sheet, are kinetically more important than those between the N- and C-terminal strands.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

