Selection bias in gene extraction on the basis of microarray gene-expression data
- PMID: 11983868
- PMCID: PMC124442
- DOI: 10.1073/pnas.102102699
Selection bias in gene extraction on the basis of microarray gene-expression data
Abstract
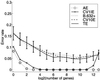
In the context of cancer diagnosis and treatment, we consider the problem of constructing an accurate prediction rule on the basis of a relatively small number of tumor tissue samples of known type containing the expression data on very many (possibly thousands) genes. Recently, results have been presented in the literature suggesting that it is possible to construct a prediction rule from only a few genes such that it has a negligible prediction error rate. However, in these results the test error or the leave-one-out cross-validated error is calculated without allowance for the selection bias. There is no allowance because the rule is either tested on tissue samples that were used in the first instance to select the genes being used in the rule or because the cross-validation of the rule is not external to the selection process; that is, gene selection is not performed in training the rule at each stage of the cross-validation process. We describe how in practice the selection bias can be assessed and corrected for by either performing a cross-validation or applying the bootstrap external to the selection process. We recommend using 10-fold rather than leave-one-out cross-validation, and concerning the bootstrap, we suggest using the so-called .632+ bootstrap error estimate designed to handle overfitted prediction rules. Using two published data sets, we demonstrate that when correction is made for the selection bias, the cross-validated error is no longer zero for a subset of only a few genes.
Figures




References
-
- Xiong M, Li W, Zhao J, Jin L, Boerwinkle E. Mol Genet Metab. 2001;73:239–247. - PubMed
-
- Dudoit S, Fridlyand J, Speed T P. J Am Stat Assoc. 2002;97:77–87.
-
- Golub T R, Slonim D K, Tamayo P, Huard C, Gassenbeck M, Mesirov J P, Coller H, Loh M L, Downing J R, Caligiuri M A, et al. Science. 1999;286:531–537. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

