A chloroplast-resident DNA methyltransferase is responsible for hypermethylation of chloroplast genes in Chlamydomonas maternal gametes
- PMID: 11983892
- PMCID: PMC122878
- DOI: 10.1073/pnas.082120199
A chloroplast-resident DNA methyltransferase is responsible for hypermethylation of chloroplast genes in Chlamydomonas maternal gametes
Abstract
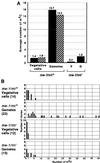
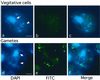
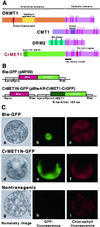
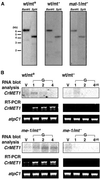
Chloroplast DNA of the green alga Chlamydomonas reinhardtii is maternally inherited. Methylation mapping directly revealed that, before mating, chloroplast DNA of maternal (mating type plus; mt(+)) gametes is heavily methylated whereas that of paternal (mating type minus; mt(-)) gametes is not. Indirect immunofluorescence analyses with anti-5-methylcytosine mAbs visually showed methylation to occur exclusively in chloroplast DNA of mt(+) gametes, and not in mt(-) gametes or nuclear DNA of either mt. To clarify the relationship between methylation and maternal inheritance of chloroplast DNA, we have isolated and characterized a cDNA encoding a DNA methyltransferase. The deduced protein, CrMET1, consists of 1,344 aa and contains a conserved catalytic domain at the C terminal and a nonconserved N-terminal region. The predicted N-terminal region has an arginine-rich domain, suggesting CrMET1 is transferred to chloroplasts. This finding could be directly shown by green fluorescent protein epifluorescence microscopy analyses. CrMET1 transcripts were found to be absent in both mt(+) and mt(-) vegetative cells. Upon gametogenesis, however, transcript levels clearly increased in mt(+) but not mt(-) cells. These experiments suggest that the CrMET1 protein is located in chloroplasts and that it specifically methylates cytosine residues of chloroplast DNA in mt(+) gametes. This conclusion was further strengthened by the observation that, during gametogenesis, CrMET1 is expressed in a mt(-) mutant, mat-1, whose chloroplast DNA is heavily methylated in gametes and paternally inherited. The results provide evidence that cytosine methylation plays a critical role in maternal inheritance of chloroplast genes in C. reinhardtii.
Figures


References
-
- Sager R. Cytoplasmic Genes and Organelles. New York: Academic; 1972.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

