Reverse engineering gene networks using singular value decomposition and robust regression
- PMID: 11983907
- PMCID: PMC122920
- DOI: 10.1073/pnas.092576199
Reverse engineering gene networks using singular value decomposition and robust regression
Abstract
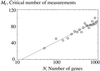
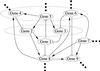
We propose a scheme to reverse-engineer gene networks on a genome-wide scale using a relatively small amount of gene expression data from microarray experiments. Our method is based on the empirical observation that such networks are typically large and sparse. It uses singular value decomposition to construct a family of candidate solutions and then uses robust regression to identify the solution with the smallest number of connections as the most likely solution. Our algorithm has O(log N) sampling complexity and O(N(4)) computational complexity. We test and validate our approach in a series of in numero experiments on model gene networks.
Figures






References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

