The evolutionary position of nematodes
- PMID: 11985779
- PMCID: PMC102755
- DOI: 10.1186/1471-2148-2-7
The evolutionary position of nematodes
Abstract
Background: The complete genomes of three animals have been sequenced by global research efforts: a nematode worm (Caenorhabditis elegans), an insect (Drosophila melanogaster), and a vertebrate (Homo sapiens). Remarkably, their relationships have yet to be clarified. The confusion concerns the enigmatic position of nematodes. Traditionally, nematodes have occupied a basal position, in part because they lack a true body cavity. However, the leading hypothesis now joins nematodes with arthropods in a molting clade, Ecdysozoa, based on data from several genes.
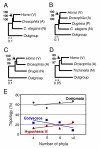
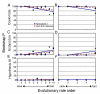
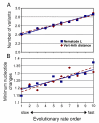
Results: We tested the Ecdysozoa hypothesis with analyses of more than 100 nuclear protein alignments, under conditions that would expose biases, and found that it was not supported. Instead, we found significant support for the traditional hypothesis, Coelomata. Our result is robust to different rates of sequence change among genes and lineages, different numbers of taxa, and different species of nematodes.
Conclusion: We conclude that insects (arthropods) are genetically and evolutionarily closer to humans than to nematode worms.
Figures
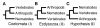
References
-
- Hyman LH. The Invertebrates. New York: McGraw-Hill; 1940.
-
- Carroll SB, Grenier JK, Weatherbee SD. In: From DNA to Diversity. Malden MA, editor. Blackwell Science; 2001.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

