Analysis of an Autographa californica multicapsid nucleopolyhedrovirus lef-6-null virus: LEF-6 is not essential for viral replication but appears to accelerate late gene transcription
- PMID: 11991978
- PMCID: PMC137020
- DOI: 10.1128/jvi.76.11.5503-5514.2002
Analysis of an Autographa californica multicapsid nucleopolyhedrovirus lef-6-null virus: LEF-6 is not essential for viral replication but appears to accelerate late gene transcription
Abstract
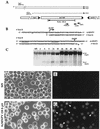
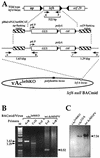
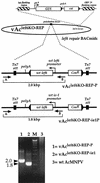
The Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV) lef-6 gene was previously shown to be necessary for optimal transcription from an AcMNPV late promoter in transient late expression assays. In the present study, we examined the expression and cellular localization of lef-6 during the AcMNPV infection cycle and generated a lef-6-null virus for studies of the role of lef-6 in the infection cycle. Transcription of lef-6 was detected from 4 to 48 h postinfection, and the LEF-6 protein was identified in dense regions of infected cell nuclei, a finding consistent with its potential role as a late transcription factor. To examine lef-6 in the context of the AcMNPV infection cycle, we deleted the lef-6 gene from an AcMNPV genome propagated as an infectious BACmid in Escherichia coli. Unexpectedly, the resulting AcMNPV lef-6-null BACmid (vAc(lef6KO)) was able to propagate in cell culture, although virus yields were substantially reduced. Thus, the lef-6 gene is not essential for viral replication in Sf9 cells. Two "repair" AcMNPV BACmids (vAc(lef6KO-REP-P) and vAc(lef6KO-REP-ie1P)) were generated by transposition of the lef-6 gene into the polyhedrin locus of the vAc(lef6KO) BACmid. Virus yields from the two repair viruses were similar to those from wild-type AcMNPV or a control (BACmid-derived) virus. The lef-6-null BACmid (vAc(lef6KO)) was further examined to determine whether the deletion of lef-6 affected DNA replication or late gene transcription in the context of an infection. The lef-6 deletion did not appear to affect viral DNA replication. Using Northern blot analysis, we found that although early transcription was apparently unaffected, both late and very late transcription were delayed in cells infected with the lef-6-null BACmid. This phenotype was rescued in viruses containing the lef-6 gene reinserted into the polyhedrin locus. Thus, the lef-6 gene was not essential for either viral DNA replication or late gene transcription, but the absence of lef-6 resulted in a substantial delay in the onset of late transcription. Therefore, lef-6 appears to accelerate the infection cycle of AcMNPV.
Figures

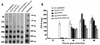

Similar articles
-
The AcMNPV pp31 gene is not essential for productive AcMNPV replication or late gene transcription but appears to increase levels of most viral transcripts.Virology. 2007 Aug 15;365(1):34-47. doi: 10.1016/j.virol.2007.02.034. Epub 2007 Apr 30. Virology. 2007. PMID: 17467768 Free PMC article.
-
Analysis of an Autographa californica nucleopolyhedrovirus lef-11 knockout: LEF-11 is essential for viral DNA replication.J Virol. 2002 Mar;76(6):2770-9. doi: 10.1128/jvi.76.6.2770-2779.2002. J Virol. 2002. PMID: 11861844 Free PMC article.
-
The Autographa californica multiple nucleopolyhedrovirus lef-5 gene is required for productive infection.Virology. 2011 Jul 20;416(1-2):54-64. doi: 10.1016/j.virol.2011.04.019. Epub 2011 May 23. Virology. 2011. PMID: 21601232
-
Involvement of host factors in transcription from baculovirus very late promoters -- a review.Gene. 1997 Apr 29;190(1):113-8. doi: 10.1016/s0378-1119(96)00827-x. Gene. 1997. PMID: 9185856 Review.
-
Baculovirus--insect cell interactions.Cytotechnology. 1996;20(1-3):73-93. doi: 10.1007/BF00350390. Cytotechnology. 1996. PMID: 8987578 Review.
Cited by
-
Use of bacterial artificial chromosomes in baculovirus research and recombinant protein expression: current trends and future perspectives.ISRN Microbiol. 2012 Sep 12;2012:628797. doi: 10.5402/2012/628797. Print 2012. ISRN Microbiol. 2012. PMID: 23762754 Free PMC article.
-
Characterization of AcMNPV with a deletion of ac68 gene.Virus Genes. 2008 Aug;37(1):119-27. doi: 10.1007/s11262-008-0238-9. Epub 2008 May 16. Virus Genes. 2008. PMID: 18483845
-
A DNA Binding Protein Is Required for Viral Replication and Transcription in Bombyx mori Nucleopolyhedrovirus.PLoS One. 2016 Jul 14;11(7):e0159149. doi: 10.1371/journal.pone.0159149. eCollection 2016. PLoS One. 2016. PMID: 27414795 Free PMC article.
-
Comparison of CRISPR-Cas9 Tools for Transcriptional Repression and Gene Disruption in the BEVS.Viruses. 2021 Sep 24;13(10):1925. doi: 10.3390/v13101925. Viruses. 2021. PMID: 34696355 Free PMC article.
-
The AcMNPV pp31 gene is not essential for productive AcMNPV replication or late gene transcription but appears to increase levels of most viral transcripts.Virology. 2007 Aug 15;365(1):34-47. doi: 10.1016/j.virol.2007.02.034. Epub 2007 Apr 30. Virology. 2007. PMID: 17467768 Free PMC article.
References
-
- Ahrens, C. H., D. J. Leisy, and G. F. Rohrmann. 1996. Baculovirus DNA replication, p. 855-872. In M. L. DePamphilis (ed.), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
-
- Ayres, M. D., S. C. Howard, J. Kuzio, M. Lopez-Ferber, and R. D. Possee. 1994. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology 202:586-605. - PubMed
-
- Bideshi, D. K., and B. A. Federici. 2000. The Trichoplusia ni granulovirus helicase is unable to support replication of Autographa californica multicapsid nucleopolyhedrovirus in cells and larvae of T. ni. J. Gen. Virol. 81: 1593-1599. - PubMed
-
- Blissard, G. W., R. L. Quant-Russell, G. F. Rohrmann, and G. S. Beaudreau. 1989. Nucleotide sequence, transcriptional mapping, and temporal expression of the gene encoding P39, a major structural protein of the multicapsid nuclear polyhedrosis virus of Orgyia pseudotsugata. Virology 168:354-362. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

