A complete sequence of the T. tengcongensis genome
- PMID: 11997336
- PMCID: PMC186588
- DOI: 10.1101/gr.219302
A complete sequence of the T. tengcongensis genome
Abstract
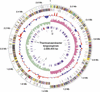
Thermoanaerobacter tengcongensis is a rod-shaped, gram-negative, anaerobic eubacterium that was isolated from a freshwater hot spring in Tengchong, China. Using a whole-genome-shotgun method, we sequenced its 2,689,445-bp genome from an isolate, MB4(T) (Genbank accession no. AE008691). The genome encodes 2588 predicted coding sequences (CDS). Among them, 1764 (68.2%) are classified according to homology to other documented proteins, and the rest, 824 CDS (31.8%), are functionally unknown. One of the interesting features of the T. tengcongensis genome is that 86.7% of its genes are encoded on the leading strand of DNA replication. Based on protein sequence similarity, the T. tengcongensis genome is most similar to that of Bacillus halodurans, a mesophilic eubacterium, among all fully sequenced prokaryotic genomes up to date. Computational analysis on genes involved in basic metabolic pathways supports the experimental discovery that T. tengcongensis metabolizes sugars as principal energy and carbon source and utilizes thiosulfate and element sulfur, but not sulfate, as electron acceptors. T. tengcongensis, as a gram-negative rod by empirical definitions (such as staining), shares many genes that are characteristics of gram-positive bacteria whereas it is missing molecular components unique to gram-negative bacteria. A strong correlation between the G + C content of tDNA and rDNA genes and the optimal growth temperature is found among the sequenced thermophiles. It is concluded that thermophiles are a biologically and phylogenetically divergent group of prokaryotes that have converged to sustain extreme environmental conditions over evolutionary timescale.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
