Single-molecule height measurements on microsomal cytochrome P450 in nanometer-scale phospholipid bilayer disks
- PMID: 11997441
- PMCID: PMC124470
- DOI: 10.1073/pnas.062565599
Single-molecule height measurements on microsomal cytochrome P450 in nanometer-scale phospholipid bilayer disks
Abstract
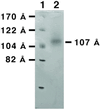
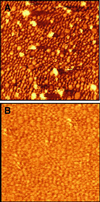
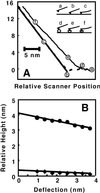
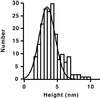
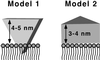
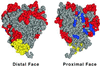
The architecture of membrane proteins in their native environment of the phospholipid bilayer is critical for understanding physiological function, but has been difficult to realize experimentally. In this communication we describe the incorporation of a membrane-anchored protein into a supported phospholipid bilayer. Cytochrome P450 2B4 solubilized and purified from the hepatic endoplasmic reticulum was incorporated into phospholipid bilayer nanostructures and oriented on a surface for visualization by atomic force microscopy. Individual P450 molecules were observed protruding from the bilayer surface. Problems associated with deformation of the protein by the atomic force microscopy probe were avoided by analyzing force-dependent height measurements to quantitate the height of the protein above the bilayer surface. Measurements of the atomic force microscopy cantilever deflection as a function of probe-sample separation reveal that the top of the P450 opposite the N-terminal membrane anchor region sits 3.5 nanometers above the phospholipid-water boundary. Models of the orientation of the enzyme are presented and discussed in relation to membrane interactions and interaction with cytochrome P450 reductase.
Figures


References
-
- Ortiz de Montellano P R. Cytochrome P-450: Structure, Mechanism, and Biochemistry. New York: Plenum; 1986.
-
- Williams P A, Cosme J, Sridhar V, Johnson E F, McRee D E. Mol Cell. 2000;5:121–131. - PubMed
-
- Strobel H W, Lu A Y, Heidema J, Coon M J. J Biol Chem. 1970;245:4851–4854. - PubMed
-
- Ingelman-Sundberg M, Haaparanta T, Rydstrom J. Biochemistry. 1981;20:4100–4106. - PubMed
-
- Ingelman-Sundberg M, Blanck J, Smettan G, Ruckpaul K. Eur J Biochem. 1983;134:157–162. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

