Nucleosome remodeling by the human SWI/SNF complex requires transient global disruption of histone-DNA interactions
- PMID: 11997502
- PMCID: PMC133810
- DOI: 10.1128/MCB.22.11.3653-3662.2002
Nucleosome remodeling by the human SWI/SNF complex requires transient global disruption of histone-DNA interactions
Abstract
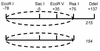
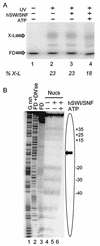
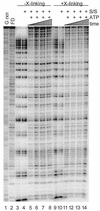
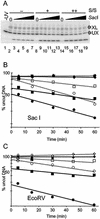
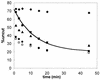
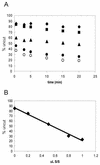
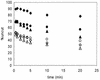
We utilized a site-specific cross-linking technique to investigate the mechanism of nucleosome remodeling by hSWI/SNF. We found that a single cross-link between H2B and DNA virtually eliminates the accumulation of stably remodeled species as measured by restriction enzyme accessibility assays. However, cross-linking the histone octamer to nucleosomal DNA does not inhibit remodeling as monitored by DNase I digestion assays. Importantly, we found that the restriction enzyme-accessible species can be efficiently cross-linked after remodeling and that the accessible state does not require continued ATP hydrolysis. These results imply that the generation of stable remodeled states requires at least transient disruption of histone-DNA interactions throughout the nucleosome, while hSWI/SNF-catalyzed disruption of just local histone-DNA interactions yields less-stable remodeled states that still display an altered DNase I cleavage pattern. The implications of these results for models of the mechanism of SWI/SNF-catalyzed nucleosome remodeling are discussed.
Figures

References
-
- Angelov, D., M. Chara, M. Seve, J. Côté, S. Khochbin, and S. Dimitrov. 2000. Differential remodeling of the HIV-1 nucleosome upon transcription activators and SWI/SNF complex binding. J. Mol. Biol. 302:315-326. - PubMed
-
- Boyer, L. A., X. Shao, R. H. Ebright, and C. L. Peterson. 2000. Roles of the histone H2A-H2B dimers and the (H3-H4)2 tetramer in nucleosome remodeling by the SWI-SNF complex. J. Biol. Chem. 275:11545-11552. - PubMed
-
- Cairns, B. R., Y. Lorch, Y. Li, M. Zhang, L. Lacomis, H. Erdjument-Bromage, P. Tempst, J. Du, B. Laurent, and R. D. Kornberg. 1996. RSC, an essential, abundant chromatin-remodeling complex. Cell 87:1249-1260. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
