Cyclic AMP blocks cell growth through Raf-1-dependent and Raf-1-independent mechanisms
- PMID: 11997508
- PMCID: PMC133826
- DOI: 10.1128/MCB.22.11.3717-3728.2002
Cyclic AMP blocks cell growth through Raf-1-dependent and Raf-1-independent mechanisms
Abstract
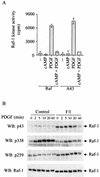
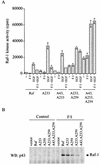
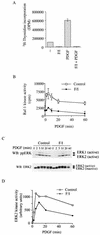
It is widely accepted that cyclic AMP (cAMP) can block cell growth by phosphorylating Raf-1 on serine 43 and inhibiting signaling to extracellular signal-regulated protein kinase. We show that the suppression of Raf-1 by cAMP is considerably more complex than previously reported. When cellular cAMP is elevated, Raf-1 is phosphorylated on three residues (S43, S233, and S259), which work independently to block Raf-1. Both Ras-dependent and Ras-independent processes are disrupted. However, when cAMP-insensitive versions of Raf-1 are expressed in NIH 3T3 cells, their growth is still strongly suppressed when cAMP is elevated. Thus, although Raf-1 appears to be an important cAMP target, other pathways are also targeted by cAMP, providing alternative mechanisms that lead to suppression of cell growth.
Figures







References
-
- Abraham, D., K. Podar, M. Pacher, M. Kubicek, N. Welzel, B. A. Hemmings, S. M. Dilworth, H. Mischak, W. Kolch, and M. Baccarini. 2000. Raf-1-associated protein phosphatase 2A as a positive regulator of kinase activation. J. Biol. Chem. 275:22300-22304. - PubMed
-
- Ally, S., T. Clair, D. Katsaros, G. Tortora, H. Yokozaki, R. A. Finch, T. L. Avery, and Y. S. Cho-Chung. 1989. Inhibition of growth and modulation of gene expression in human lung carcinoma in athymic mice by site-selective 8-Cl-cyclic adenosine monophosphate. Cancer Res. 49:5650-5655. - PubMed
-
- Avruch, J., A. Khokhlatchev, J. M. Kyriakis, Z. Luo, G. Tzivion, D. Vavvas, and X. F. Zhang. 2001. Ras activation of the Raf kinase: tyrosine kinase recruitment of the MAP kinase cascade. Recent Prog. Horm. Res. 56:127-155. - PubMed
-
- Bos, J. L., J. de Rooij, and K. A. Reedquist. 2001. Rap1 signalling: adhering to new models. Nat. Rev. Mol. Cell Biol. 2:369-377. - PubMed
-
- Boyle, W. J., P. van der Geer, and T. Hunter. 1991. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 201:110-149. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
