Applications of quantitative PCR in the biosafety and genetic stability assessment of biotechnology products
- PMID: 11999695
- PMCID: PMC7148957
- DOI: 10.1016/s1389-0352(01)00043-5
Applications of quantitative PCR in the biosafety and genetic stability assessment of biotechnology products
Abstract
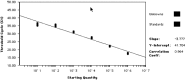
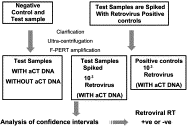
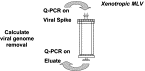
High throughput screening, increased accuracy and the coupling of real-time quantitative PCR (Q-PCR) to robotic set-up systems are beginning to revolutionise biotechnology. Applications of Q-PCR within biotechnology are discussed with particular emphasis on the following areas of biosafety and genetic stability testing: (a) determination of the biodistribution of gene therapy vectors in animals; (b) quantification of the residual DNA in final product therapeutics; (c) detection of viral and bacterial nucleic acid in contaminated cell banks and final products; (d) quantification of the level of virus removal in process validation viral clearance studies; (e) specific detection of retroviral RT activity in vaccines with high sensitivity; and (f) transgene copy number determination for monitoring genetic stability during production. Methods employed for Q-PCR assay validation as required in ICH Topic Q2A Validation of Analytical Methods: Definitions and Terminology (1st June 1995) are also reviewed.
Figures


References
-
- Aberham C., Pendl C., Gros P., Zerlauth G., Gessner M. A quantitative, internally controlled real-time PCR assay for the detection of parvovirus B19 DNA. J. Virol. Methods. 2001;92:183–192. - PubMed
-
- Allan G.M., Ellis J.A. Porcine circoviruses: a review. J. Vet. Diagn. Invest. 2000;12:3–14. - PubMed
-
- Arnold B.A., Hepler R.W., Keller P.M. One step fluorescent probe product enhanced reverse transcriptase assay. Biotechniques. 1998;25:98–106. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

