Telomere measurement by quantitative PCR
- PMID: 12000852
- PMCID: PMC115301
- DOI: 10.1093/nar/30.10.e47
Telomere measurement by quantitative PCR
Abstract
It has long been presumed impossible to measure telomeres in vertebrate DNA by PCR amplification with oligonucleotide primers designed to hybridize to the TTAGGG and CCCTAA repeats, because only primer dimer-derived products are expected. Here we present a primer pair that eliminates this problem, allowing simple and rapid measurement of telomeres in a closed tube, fluorescence-based assay. This assay will facilitate investigations of the biology of telomeres and the roles they play in the molecular pathophysiology of diseases and aging.
Figures

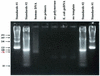
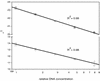

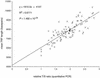
Similar articles
-
Real-time PCR assay for measurement of mouse telomeres.Comp Med. 2006 Feb;56(1):17-22. Comp Med. 2006. PMID: 16521855
-
Analysis of telomeres and telomerase.Curr Protoc Cell Biol. 2003 Nov;Chapter 18:Unit 18.6. doi: 10.1002/0471143030.cb1806s20. Curr Protoc Cell Biol. 2003. PMID: 18228424
-
Estimation of the amount of telomere molecules in different human age groups and the telomere increasing effect of acupuncture and shiatsu on St.36, using synthesized basic units of the human telomere molecules as reference control substances for the bi-digital O-ring test resonance phenomenon.Acupunct Electrother Res. 1998;23(3-4):185-206. doi: 10.3727/036012998816356472. Acupunct Electrother Res. 1998. PMID: 10193703
-
New developments in telomere length analysis.Exp Gerontol. 2005 May;40(5):363-8. doi: 10.1016/j.exger.2005.02.008. Epub 2005 Mar 29. Exp Gerontol. 2005. PMID: 15919587 Review.
-
The polymerase chain reaction: new variations on an old theme.Curr Opin Biotechnol. 1995 Feb;6(1):24-9. doi: 10.1016/0958-1669(95)80005-0. Curr Opin Biotechnol. 1995. PMID: 7894078 Review.
Cited by
-
DNA-methylation-based telomere length estimator: comparisons with measurements from flow FISH and qPCR.Aging (Albany NY). 2021 Jun 3;13(11):14675-14686. doi: 10.18632/aging.203126. Epub 2021 Jun 3. Aging (Albany NY). 2021. PMID: 34083495 Free PMC article.
-
The association between leukocyte telomere lengths and sleep instability based on cardiopulmonary coupling analysis.Sleep Breath. 2015 Sep;19(3):963-8. doi: 10.1007/s11325-014-1110-x. Epub 2015 Jan 28. Sleep Breath. 2015. PMID: 25628010
-
Eroded human telomeres are more prone to remain uncapped and to trigger a G2 checkpoint response.Nucleic Acids Res. 2013 Jan;41(2):900-11. doi: 10.1093/nar/gks1121. Epub 2012 Nov 27. Nucleic Acids Res. 2013. PMID: 23193277 Free PMC article.
-
Parental responsiveness moderates the association between early-life stress and reduced telomere length.Dev Psychopathol. 2013 Aug;25(3):577-585. doi: 10.1017/S0954579413000011. Epub 2013 Mar 26. Dev Psychopathol. 2013. PMID: 23527512 Free PMC article.
-
Early exclusive breastfeeding is associated with longer telomeres in Latino preschool children.Am J Clin Nutr. 2016 Aug;104(2):397-405. doi: 10.3945/ajcn.115.115428. Epub 2016 Jul 20. Am J Clin Nutr. 2016. PMID: 27440083 Free PMC article.
References
-
- Lansdorp P.M., Verwoerd,N.P., van de Rijke,F.M., Dragowska,V., Little,M.T., Dirks,R.W., Raap,A.K. and Tanke,H.J. (1996) Heterogeneity in telomere length of human chromosomes. Hum. Mol. Genet., 5, 685–691. - PubMed
-
- Bryant J.E., Hutchings,K.G., Moyzis,R.K. and Griffith,J.K. (1997) Measurement of telomeric DNA content in human tissues. Biotechniques, 23, 476–478, 480, 482, passim. - PubMed
-
- Norwood D. and Dimitrov,D.S. (1998) Sensitive method for measuring telomere lengths by quantifying telomeric DNA content of whole cells. Biotechniques, 25, 1040–1045. - PubMed
-
- Nakamura Y., Hirose,M., Matsuo,H., Tsuyama,N., Kamisango,K. and Ide,T. (1999) Simple, rapid, quantitative and sensitive detection of telomere repeats in cell lysate by a hybridization protection assay. Clin. Chem., 45, 1718–1724. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

