Plasmid-encoded asp operon confers a proton motive metabolic cycle catalyzed by an aspartate-alanine exchange reaction
- PMID: 12003930
- PMCID: PMC135062
- DOI: 10.1128/JB.184.11.2906-2913.2002
Plasmid-encoded asp operon confers a proton motive metabolic cycle catalyzed by an aspartate-alanine exchange reaction
Abstract
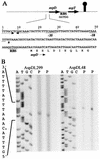
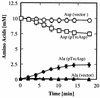
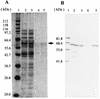
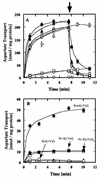
Tetragenococcus halophila D10 catalyzes the decarboxylation of L-aspartate with nearly stoichiometric release of L-alanine and CO(2). This trait is encoded on a 25-kb plasmid, pD1. We found in this plasmid a putative asp operon consisting of two genes, which we designated aspD and aspT, encoding an L-aspartate-beta-decarboxylase (AspD) and an aspartate-alanine antiporter (AspT), respectively, and determined the nucleotide sequences. The sequence analysis revealed that the genes of the asp operon in pD1 were in the following order: promoter --> aspD --> aspT. The deduced amino acid sequence of AspD showed similarity to the sequences of two known L-aspartate-beta-decarboxylases from Pseudomonas dacunhae and Alcaligenes faecalis. Hydropathy analyses suggested that the aspT gene product encodes a hydrophobic protein with multiple membrane-spanning regions. The operon was subcloned into the Escherichia coli expression vector pTrc99A, and the two genes were cotranscribed in the resulting plasmid, pTrcAsp. Expression of the asp operon in E. coli coincided with appearance of the capacity to catalyze the decarboxylation of aspartate to alanine. Histidine-tagged AspD (AspDHis) was also expressed in E. coli and purified from cell extracts. The purified AspDHis clearly exhibited activity of L-aspartate-beta-decarboxylase. Recombinant AspT was solubilized from E. coli membranes and reconstituted in proteoliposomes. The reconstituted AspT catalyzed self-exchange of aspartate and electrogenic heterologous exchange of aspartate with alanine. Thus, the asp operon confers a proton motive metabolic cycle consisting of the electrogenic aspartate-alanine antiporter and the aspartate decarboxylase, which keeps intracellular levels of alanine, the countersubstrate for aspartate, high.
Figures

References
-
- Abe, K., H. Hayashi, and P. C. Maloney. 1996. Exchange of aspartate and alanine. J. Biol. Chem. 271:3079-3084. - PubMed
-
- Abe, K., Z. S. Ruan, and P. C. Maloney. 1996. Cloning, sequencing, and expression in Escherichia coli of OxlT, the oxalate:formate exchange protein of Oxalobacter formigenes. J. Biol. Chem. 271:6789-6793. - PubMed
-
- Ambudkar, S. V., and P. C. Maloney. 1986. Bacterial anion exchange, use of osmolyte during solubilization and reconstitution of phosphate-linked antiport from Streptococcus lactis. J. Biol. Chem. 261:10079-10086. - PubMed
-
- Anantharam, V., M. J. Allison, and P. C. Maloney. 1989. Oxalate:formate exchange. The basis for energy coupling in Oxalobacter. J. Biol. Chem. 264:7244-7250. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

