CLOUDS, a protocol for deriving a molecular proton density via NMR
- PMID: 12011433
- PMCID: PMC124467
- DOI: 10.1073/pnas.082114199
CLOUDS, a protocol for deriving a molecular proton density via NMR
Erratum in
- Proc Natl Acad Sci U S A 2002 Aug 6;99(16):10941
Abstract
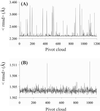
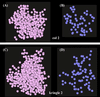
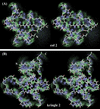
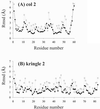
We demonstrate the feasibility of computing realistic spatial proton distributions for proteins in solution from experimental NMR nuclear Overhauser effect data only and with minimal assignments. The method, CLOUDS, relies on precise and abundant interproton distance restraints calculated via a relaxation matrix analysis of sets of experimental nuclear Overhauser effect spectroscopy crosspeaks. The MIDGE protocol was adapted for this purpose. A gas of unassigned, unconnected H atoms is condensed into a structured proton distribution (cloud) via a molecular dynamics simulated-annealing scheme in which the internuclear distances and van der Waals repulsive terms are the only active restraints. Proton densities are generated by combining a large number of such clouds, each computed from a different trajectory. After filtering by reference to the cloud closest to the mean, a minimal dispersion proton density (foc) is identified. The latter affords a quasi-continuous hydrogen-only probability distribution that conveys immediate information on the protein surface topology (grooves, protrusions, potential binding site cavities, etc.), directly related to the molecular structure. Feasibility of the method was tested on NMR data measured on two globular protein domains of low regular secondary structure content, the col 2 domain of human matrix metalloproteinase-2 and the kringle 2 domain of human plasminogen, of 60 and 83 amino acid residues, respectively.
Figures



References
-
- Boelens R, Koning T M G, Van Der Marel G A, Van Boom J H, Kaptein R. J Magn Reson. 1989;82:290–308.
-
- Borgias B A, James T L. J Magn Reson. 1990;87:475–487.
-
- Zhang Q, Chen J Y, Gozansky E K, Zhu F, Jackson P L, Gorenstein D G. J Magn Reson B. 1995;106:164–169. - PubMed
-
- Madrid M, Llinás E, Llinás M. J Magn Reson. 1991;93:329–346.
-
- Wüthrich K. NMR of Proteins and Nucleic Acids. New York: Wiley; 1986. pp. 117–199.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

