The root of the angiosperms revisited
- PMID: 12011443
- PMCID: PMC124492
- DOI: 10.1073/pnas.092136399
The root of the angiosperms revisited
Abstract
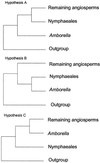
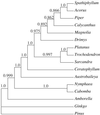
Most recent phylogenetic analyses of basal angiosperms have converged on the placement of Amborella as sister to all other extant angiosperms. However, certain recent studies suggest that Amborella and Nymphaeales (water lilies) form a clade sister to all remaining angiosperms or that Nymphaeales alone are the sister to the remaining angiosperms. We report here (i) maximum parsimony, maximum likelihood, and Bayesian phylogenetic analyses of 11 genes (>15,000 bp per taxon) for 16 taxa, (ii) maximum parsimony analysis for a subset of these genes for 104 taxa, and (iii) tests of alternative rootings with the nonparametric bootstrap and the likelihood ratio test with the parametric bootstrap. In addition, we use simulation analyses to examine the amount of bias that may be present in our methods of phylogeny estimation. Amborella continues to receive strong bootstrap support as the sister to all other extant angiosperms, and three of four tests reject alternative hypotheses of the angiosperm root. Although we cannot conclusively choose between Amborella vs. Amborella + Nymphaeales as sister to all other angiosperms, most analyses favor the former rooting.
Figures


References
-
- Mathews S, Donoghue M J. Science. 1999;286:947–950. - PubMed
-
- Parkinson C L, Adams K L, Palmer J D. Curr Biol. 1999;9:1485–1488. - PubMed
-
- Qiu Y-L, Lee J, Bernasconi-Quadroni F, Soltis D E, Soltis P S, Zanis M, Zimmer E A, Chen Z, Savolainen V, Chase M W. Nature (London) 1999;402:404–407. - PubMed
-
- Soltis P S, Soltis D E, Chase M W. Nature (London) 1999;402:402–404. - PubMed
-
- Soltis D E, Soltis P S, Chase M W, Mort M E, Albach D C, Zanis M, Savolainen V, Hahn W H, Hoot S B, Fay M F, et al. Bot J Linn Soc. 2000;133:381–461.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

