CAU-1, a subclass B3 metallo-beta-lactamase of low substrate affinity encoded by an ortholog present in the Caulobacter crescentus chromosome
- PMID: 12019096
- PMCID: PMC127251
- DOI: 10.1128/AAC.46.6.1823-1830.2002
CAU-1, a subclass B3 metallo-beta-lactamase of low substrate affinity encoded by an ortholog present in the Caulobacter crescentus chromosome
Abstract
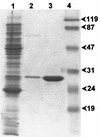
The sequenced chromosome of Caulobacter crescentus CB15 encodes a hypothetical protein that exhibits significant similarity (30 to 35% identical residues) to metallo-beta-lactamases of subclass B3. An allelic variant of this gene (divergent by 3% of its nucleotides) was cloned in Escherichia coli from C. crescentus type strain DSM4727. Expression studies confirmed the metallo-beta-lactamase activity of its product, CAU-1. The enzyme produced in E. coli was purified by two ion-exchange chromatography steps. CAU-1 contains a 29-kDa polypeptide with an alkaline isoelectric pH (> 9), and unlike the L1 enzyme of Stenotrophomonas maltophilia, the native form is monomeric. Kinetic analysis revealed a preferential activity toward penicillins, carbapenems, and narrow-spectrum cephalosporins, while oxyimino cephalosporins were poorly or not hydrolyzed. Affinities for the various beta-lactams were poor overall (K(m) values were always > 100 microM and often > 400 microM). The interaction with divalent ion chelators appeared to occur by a mechanism similar to that prevailing in other members of subclass B3. In C. crescentus, the CAU-1 enzyme is produced independently of beta-lactam exposure and, interestingly, the bla(CAU) determinant is bracketed by three other genes, including two genes encoding enzymes involved in methionine biosynthesis and a gene encoding a putative transcriptional regulator, in an operon-like structure. The CAU-1 enzyme is the first example of a metallo-beta-lactamase in a member of the alpha subdivision of the class Proteobacteria:
Figures



References
-
- Ambler, R. P. 1980. The structure of β-lactamases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 289:321-331. - PubMed
-
- Bellais, S., S. Léotard, L. Poirel, T. Naas, and P. Nordmann. 1999. Molecular characterization of a carbapenem-hydrolyzing β-lactamase from Chryseobacterium (Flavobacterium) indologenes. FEMS Microbiol. Lett. 171:127-132. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

