Expression cloning and characterization of the C28 acyltransferase of lipid A biosynthesis in Rhizobium leguminosarum
- PMID: 12019272
- PMCID: PMC2556286
- DOI: 10.1074/jbc.M204525200
Expression cloning and characterization of the C28 acyltransferase of lipid A biosynthesis in Rhizobium leguminosarum
Abstract
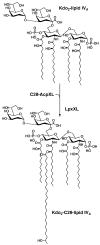
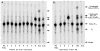
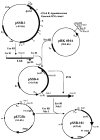
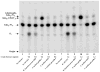
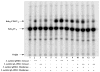
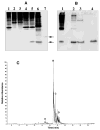
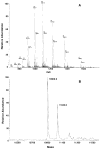
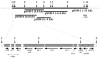
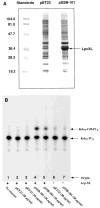
An unusual feature of lipid A from plant endosymbionts of the Rhizobiaceae family is the presence of a 27-hydroxyoctacosanoic acid (C28) moiety. An enzyme that incorporates this acyl chain is present in extracts of Rhizobium leguminosarum, Rhizobium etli, and Sinorhizobium meliloti but not Escherichia coli. The enzyme transfers 27-hydroxyoctacosanate from a specialized acyl carrier protein (AcpXL) to the precursor Kdo2 ((3-deoxy-d-manno-octulosonic acid)2)-lipid IV(A). We now report the identification of five hybrid cosmids that direct the overexpression of this activity by screening approximately 4000 lysates of individual colonies of an R. leguminosarum 3841 genomic DNA library in the host strain S. meliloti 1021. In these heterologous constructs, both the C28 acyltransferase and C28-AcpXL are overproduced. Sequencing of a 9-kb insert from cosmid pSSB-1, which is also present in the other cosmids, shows that acpXL and the lipid A acyltransferase gene (lpxXL) are close to each other but not contiguous. Nine other open reading frames around lpxXL were also sequenced. Four of them encode orthologues of fatty acid and/or polyketide biosynthetic enzymes. AcpXL purified from S. meliloti expressing pSSB-1 is fully acylated, mainly with 27-hydroxyoctacosanoate. Expression of lpxXL in E. coli behind a T7 promoter results in overproduction in vitro of the expected R. leguminosarum acyltransferase, which is C28-AcpXL-dependent and utilizes (3-deoxy-d-manno-octulosonic acid)2-lipid IV(A) as the acceptor. These findings confirm that lpxXL is the structural gene for the C28 acyltransferase. LpxXL is distantly related to the lauroyltransferase (LpxL) of E. coli lipid A biosynthesis, but highly significant LpxXL orthologues are present in Agrobacterium tumefaciens, Brucella melitensis, and all sequenced strains of Rhizobium, consistent with the occurrence of long secondary acyl chains in the lipid A molecules of these organisms.
Figures

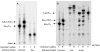

References
-
- Bhat UR, Forsberg LS, Carlson RW. J Biol Chem. 1994;269:14402–14410. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

