Distinct requirement for two stages of protein-primed initiation of reverse transcription in hepadnaviruses
- PMID: 12021318
- PMCID: PMC136195
- DOI: 10.1128/jvi.76.12.5857-5865.2002
Distinct requirement for two stages of protein-primed initiation of reverse transcription in hepadnaviruses
Abstract
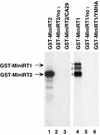
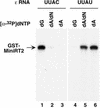
Reverse transcription in hepadnaviruses is primed by the viral reverse transcriptase (RT) (protein priming) and requires the specific interaction between the RT and a viral RNA signal termed epsilon, which bears the specific template sequence for protein priming. The product of protein priming is a short oligodeoxynucleotide which represents the 5' end of the viral minus-strand DNA and is covalently attached to the RT. We have now identified truncated RT variants from the duck hepatitis B virus that were fully active in the initial step of protein priming, i.e., the covalent attachment of the first nucleotide to the protein (RT deoxynucleotidylation), but defective in any subsequent DNA polymerization. A short sequence in the RT domain was localized that was dispensable for RT deoxynucleotidylation but essential for the subsequent DNA polymerization. These results have thus revealed two distinct stages of protein priming, i.e., the initial attachment of the first nucleotide to the RT (RT deoxynucleotidylation or initiation of protein priming) and the subsequent DNA synthesis (polymerization) to complete protein priming, with the second step entailing additional RT sequences. Two models are proposed to explain the observed differential sequence requirement for the two distinct stages of the protein priming reaction.
Figures





Similar articles
-
TP-RT domain interactions of duck hepatitis B virus reverse transcriptase in cis and in trans during protein-primed initiation of DNA synthesis in vitro.J Virol. 2012 Jun;86(12):6522-36. doi: 10.1128/JVI.00086-12. Epub 2012 Apr 18. J Virol. 2012. PMID: 22514346 Free PMC article.
-
Functional and structural dynamics of hepadnavirus reverse transcriptase during protein-primed initiation of reverse transcription: effects of metal ions.J Virol. 2008 Jun;82(12):5703-14. doi: 10.1128/JVI.02760-07. Epub 2008 Apr 9. J Virol. 2008. PMID: 18400846 Free PMC article.
-
Heat shock protein 90-independent activation of truncated hepadnavirus reverse transcriptase.J Virol. 2003 Apr;77(8):4471-80. doi: 10.1128/jvi.77.8.4471-4480.2003. J Virol. 2003. PMID: 12663754 Free PMC article.
-
Why are hepadnaviruses DNA and not RNA viruses?Trends Microbiol. 1997 Nov;5(11):447-50. doi: 10.1016/s0966-842x(97)01141-4. Trends Microbiol. 1997. PMID: 9402701 Review.
-
Hepatitis B viruses: reverse transcription a different way.Virus Res. 2008 Jun;134(1-2):235-49. doi: 10.1016/j.virusres.2007.12.024. Epub 2008 Mar 12. Virus Res. 2008. PMID: 18339439 Review.
Cited by
-
TP-RT domain interactions of duck hepatitis B virus reverse transcriptase in cis and in trans during protein-primed initiation of DNA synthesis in vitro.J Virol. 2012 Jun;86(12):6522-36. doi: 10.1128/JVI.00086-12. Epub 2012 Apr 18. J Virol. 2012. PMID: 22514346 Free PMC article.
-
Hepatitis B virus reverse transcriptase and epsilon RNA sequences required for specific interaction in vitro.J Virol. 2006 Mar;80(5):2141-50. doi: 10.1128/JVI.80.5.2141-2150.2006. J Virol. 2006. PMID: 16474122 Free PMC article.
-
Cryptic protein priming sites in two different domains of duck hepatitis B virus reverse transcriptase for initiating DNA synthesis in vitro.J Virol. 2011 Aug;85(15):7754-65. doi: 10.1128/JVI.00483-11. Epub 2011 May 18. J Virol. 2011. PMID: 21593164 Free PMC article.
-
Functional and structural dynamics of hepadnavirus reverse transcriptase during protein-primed initiation of reverse transcription: effects of metal ions.J Virol. 2008 Jun;82(12):5703-14. doi: 10.1128/JVI.02760-07. Epub 2008 Apr 9. J Virol. 2008. PMID: 18400846 Free PMC article.
-
Hepatitis B Virus Epsilon (ε) RNA Element: Dynamic Regulator of Viral Replication and Attractive Therapeutic Target.Viruses. 2023 Sep 12;15(9):1913. doi: 10.3390/v15091913. Viruses. 2023. PMID: 37766319 Free PMC article. Review.
References
-
- Allen, M. I., M. Deslauriers, C. W. Andrews, G. A. Tipples, K. A. Walters, D. L. Tyrrell, N. Brown, L. D. Condreay, et al. 1998. Identification and characterization of mutations in hepatitis B virus resistant to lamivudine. Hepatology 27:1670-1677. - PubMed
-
- Bartenschlager, R., C. Kuhn, and H. Schaller. 1992. Expression of the P-protein of the human hepatitis B virus in a vaccinia virus system and detection of the nucleocapsid-associated P-gene product by radiolabelling at newly introduced phosphorylation sites. Nucleic Acids Res. 20:195-202. - PMC - PubMed
-
- Brenkman, A. B., M. R. Heideman, V. Truniger, M. Salas, and P. C. van der Vliet. 2001. The (I/Y)XGG motif of adenovirus DNA polymerase affects template DNA binding and the transition from initiation to elongation. J. Biol. Chem. 276:29846-29853. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

