Coalescence of the sites of cowpea mosaic virus RNA replication into a cytopathic structure
- PMID: 12021357
- PMCID: PMC136224
- DOI: 10.1128/jvi.76.12.6235-6243.2002
Coalescence of the sites of cowpea mosaic virus RNA replication into a cytopathic structure
Abstract
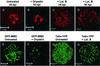
Cowpea mosaic virus (CPMV) replication induces an extensive proliferation of endoplasmic reticulum (ER) membranes, leading to the formation of small membranous vesicles where viral RNA replication takes place. Using fluorescent in situ hybridization, we found that early in the infection of cowpea protoplasts, CPMV plus-strand RNA accumulates at numerous distinct subcellular sites distributed randomly throughout the cytoplasm which rapidly coalesce into a large body located in the center of the cell, often near the nucleus. The combined use of immunostaining and a green fluorescent protein ER marker revealed that during the course of an infection, CPMV RNA colocalizes with the 110-kDa viral polymerase and other replication proteins and is always found in close association with proliferated ER membranes, indicating that these sites correspond to the membranous site of viral replication. Experiments with the cytoskeleton inhibitors oryzalin and latrunculin B point to a role of actin and not tubulin in establishing the large central structure. The induction of ER membrane proliferations in CPMV-infected protoplasts did not coincide with increased levels of BiP mRNA, indicating that the unfolded-protein response is not involved in this process.
Figures





References
-
- Assink, A. M., H. Swaans, and A. Van Kammen. 1973. The localization of virus-specific double-stranded RNA of cowpea mosaic virus in subcellular fractions of infected Vigna leaves. Virology 53:384-391. - PubMed
-
- Baluska, F., J. Jasik, H. G. Edelmann, T. Salajova, and D. Volkmann. 2001. Latrunculin B-induced plant dwarfism: plant cell elongation is F-actin-dependent. Dev. Biol. 231:113-124. - PubMed
-
- Baskin, T. I., J. E. Wilson, A. Cork, and R. E. Williamson. 1994. Morphology and microtubule organization in Arabidopsis roots exposed to oryzalin or taxol. Plant Cell Physiol. 35:935-942. - PubMed
-
- Beachy, R. N., and M. Heinlein. 2000. Role of P30 in replication and spread of TMV. Traffic 1:540-544. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

