Cowpea mosaic virus 32- and 60-kilodalton replication proteins target and change the morphology of endoplasmic reticulum membranes
- PMID: 12021362
- PMCID: PMC136232
- DOI: 10.1128/jvi.76.12.6293-6301.2002
Cowpea mosaic virus 32- and 60-kilodalton replication proteins target and change the morphology of endoplasmic reticulum membranes
Abstract
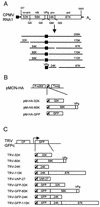
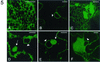
Cowpea mosaic virus (CPMV) replicates in close association with small membranous vesicles that are formed by rearrangements of intracellular membranes. To determine which of the viral proteins are responsible for the rearrangements of membranes and the attachment of the replication complex, we have expressed individual CPMV proteins encoded by RNA1 in cowpea protoplasts by transient expression and in Nicotiana benthamiana plants by using the tobacco rattle virus (TRV) expression vector. The 32-kDa protein (32K) and 60K, when expressed individually, accumulate in only low amounts but are found associated with membranes mainly derived from the endoplasmic reticulum (ER). 24K and 110K are freely soluble and accumulate to high levels. With the TRV vector, expression of 32K and 60K results in rearrangement of ER membranes. Besides, expression of 32K and 60K results in necrosis of the inoculated N. benthamiana leaves, suggesting that 32K and 60K are cytotoxic proteins. On the other hand, during CPMV infection 32K and 60K accumulate to high levels without causing necrosis.
Figures

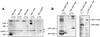


References
-
- Aldabe, R., A. Barco, and L. Carrasco. 1996. Membrane permeabilization by poliovirus proteins 2B and 2BC. J. Biol. Chem. 271:23134-23137. - PubMed
-
- Aldabe, R., and L. Carrasco. 1995. Induction of membrane proliferation by poliovirus proteins 2C and 2BC. Biochem. Biophys. Res. Commun. 206:64-76. - PubMed
-
- Bertens, P., J. Wellink, R. Goldbach, and A. van Kammen. 2000. Mutational analysis of the cowpea mosaic virus movement protein. Virology 267:199-208. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

