Deubiquitinating function of adenovirus proteinase
- PMID: 12021365
- PMCID: PMC136223
- DOI: 10.1128/jvi.76.12.6323-6331.2002
Deubiquitinating function of adenovirus proteinase
Abstract
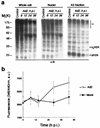
The invasion strategy of many viruses involves the synthesis of viral gene products that mimic the functions of the cellular proteins and thus interfere with the key cellular processes. Here we show that adenovirus infection is accompanied by an increased ubiquitin-cleaving (deubiquitinating) activity in the host cells. Affinity chromatography on ubiquitin aldehyde (Ubal), which was designed to identify the deubiquitinating proteases, revealed the presence of adenovirus L3 23K proteinase (Avp) in the eluate from adenovirus-infected cells. This proteinase is known to be necessary for the processing of viral precursor proteins during virion maturation. We show here that in vivo Avp deubiquitinates a number of cellular proteins. Analysis of the substrate specificity of Avp in vitro demonstrated that the protein deubiquitination by this enzyme could be as efficient as proteolytic processing of viral proteins. The structural model of the Ubal-Avp interaction revealed some similarity between S1-S4 substrate binding sites of Avp and ubiquitin hydrolases. These results may reflect the acquisition of an advantageous property by adenovirus and may indicate the importance of ubiquitin pathways in viral infection.
Figures
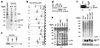


References
-
- Anderson, C. W. 1990. The proteinase polypeptide of adenovirus serotype 2 virions. Virology 177:259-272. - PubMed
-
- Andres, G., A. Alejo, C. Simon-Mateo, and M. L. Salas. 2001. African swine fever virus protease, a new viral member of the SUMO-1-specific protease family. J. Biol. Chem. 276:780-787. - PubMed
-
- Bertolaet, B. L., D. J. Clarke, M. Wolff, M. H. Watson, M. Henze, G. Divita, and S. I. Reed. 2001. UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nat. Struct. Biol. 8:417-422. - PubMed
-
- CCP4. 1994. The CCP4 suite: programs for protein crystallography. Acta Cryst. D 50:760-763. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

