Epitope mapping of a monoclonal antibody against human thrombin by H/D-exchange mass spectrometry reveals selection of a diverse sequence in a highly conserved protein
- PMID: 12021429
- PMCID: PMC2373625
- DOI: 10.1110/ps.4670102
Epitope mapping of a monoclonal antibody against human thrombin by H/D-exchange mass spectrometry reveals selection of a diverse sequence in a highly conserved protein
Abstract
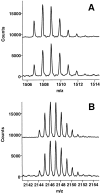
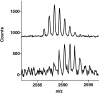
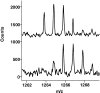
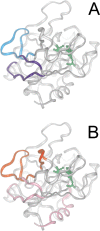
The epitope of a monoclonal antibody raised against human thrombin has been determined by hydrogen/deuterium exchange coupled to MALDI mass spectrometry. The antibody epitope was identified as the surface of thrombin that retained deuterium in the presence of the monoclonal antibody compared to control experiments in its absence. Covalent attachment of the antibody to protein G beads and efficient elution of the antigen after deuterium exchange afforded the analysis of all possible epitopes in a single MALDI mass spectrum. The epitope, which was discontinuous, consisting of two peptides close to anion-binding exosite I, was readily identified. The epitope overlapped with, but was not identical to, the thrombomodulin binding site, consistent with inhibition studies. The antibody bound specifically to human thrombin and not to murine or bovine thrombin, although these proteins share 86% identity with the human protein. Interestingly, the epitope turned out to be the more structured of two surface regions in which higher sequence variation between the three species is seen.
Figures




References
-
- Baerga-Ortiz, A., Rezaie, A.R., and Komives, E.A. 2000. Electrostatic dependence of the thrombin–thrombomodulin interaction. J. Mol. Biol. 296 651–658. - PubMed
-
- Burritt, J., DeLeo, F., McDonald, C., Prigge, J., Dinauer, M., Nakamura, M., Nauseef, W., and Jesaitis, A. 2001. Phage display epitope mapping of human neutrophil flavocytochrome b558. J. Biol. Chem. 276 2053–2061. - PubMed
-
- Chambers, G., Lawrie, L., Cash, P., and Murray, G. 2000. Proteomics: A new approach to the study of disease. J. Pathol. 192 280–288. - PubMed
-
- Dawes, J., James, K., Micklem, L., Pepper, D., and Prowse, C. 1984. Monoclonal antibodies directed against human α-thrombin and the thrombin–antithrombin III complex. Throm. Res. 36 397–409. - PubMed
-
- Fiedler, W., Borchers, C., Macht, M., Deininger, S., and Przybylski, M. 1998. Molecular characterization of a conformational epitope of hen egg white lysozyme by differential chemical modification of immune complexes and mass spectrometric peptide mapping. Bioconjug. Chem. 9 236–241. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

