An improved hydrogen bond potential: impact on medium resolution protein structures
- PMID: 12021440
- PMCID: PMC2373622
- DOI: 10.1110/ps.4890102
An improved hydrogen bond potential: impact on medium resolution protein structures
Abstract
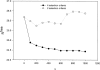
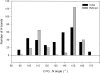
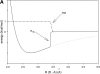
A new semi-empirical force field has been developed to describe hydrogen-bonding interactions with a directional component. The hydrogen bond potential supports two alternative target angles, motivated by the observation that carbonyl hydrogen bond acceptor angles have a bimodal distribution. It has been implemented as a module for a macromolecular refinement package to be combined with other force field terms in the stereochemically restrained refinement of macromolecules. The parameters for the hydrogen bond potential were optimized to best fit crystallographic data from a number of protein structures. Refinement of medium-resolution structures with this additional restraint leads to improved structure, reducing both the free R-factor and over-fitting. However, the improvement is seen only when stringent hydrogen bond selection criteria are used. These findings highlight common misconceptions about hydrogen bonding in proteins, and provide explanations for why the explicit hydrogen bonding terms of some popular force field sets are often best switched off.
Figures









References
-
- Avila-Sakar, A.J., Creutz, C.E., and Kretsinger, R.H. 1998. Crystal structure of bovine annexin VI in a calcium-bound state. Biochim. Biophys. Acta 1387 103–116. - PubMed
-
- Baker, E.N. and Hubbard, R.E. 1984. Hydrogen bonding in globular proteins. Prog. Biophys. Mol. Biol. 44 97–179. - PubMed
-
- Bordo, D. and Argos, P. 1994. The role of side-chain hydrogen bonds in the formation and stabilization of secondary structure in soluble proteins. J. Mol. Biol. 243 504–519. - PubMed
-
- Bowie, J.U. 1997. Helix packing angle preferences. Nat. Struct. Biol. 4 915–917. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

