Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis
- PMID: 12023303
- PMCID: PMC186273
- DOI: 10.1101/gad.969702
Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis
Abstract
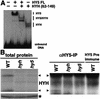
Arabidopsis COP1 acts to repress photomorphogenesis in the absence of light. It was shown that in the dark, COP1 directly interacts with the bZIP transcription factor HY5, a positive regulator of photomorphogenesis, and promotes its proteasome-mediated degradation. Here we identify a novel bZIP protein HYH, as a new target of COP1. We identify a physical and genetic interaction between HYH and COP1 and show that this interaction results in dark-specific degradation of HYH. Genetic analysis indicates that HYH is predominantly involved in blue-light regulation of development and gene expression, and that the function of HYH in part overlaps with that of HY5. The accumulation of HYH protein, not the mRNA, is dependent on the presence of HY5. Our data suggest that HYH and HY5 can, respectively, act as heterodimers and homodimers, thus mediating light-regulated expression of overlapping as well as distinct target genes. We propose that COP1 mediates light control of gene expression through targeted degradation of multiple photomorphogenesis-promoting transcription factors in the nucleus.
Figures

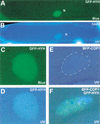


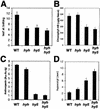
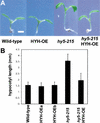
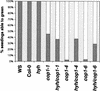

References
-
- Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng XW. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell. 1998;1:213–222. - PubMed
-
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
