Snf1 protein kinase and the repressors Nrg1 and Nrg2 regulate FLO11, haploid invasive growth, and diploid pseudohyphal differentiation
- PMID: 12024013
- PMCID: PMC133850
- DOI: 10.1128/MCB.22.12.3994-4000.2002
Snf1 protein kinase and the repressors Nrg1 and Nrg2 regulate FLO11, haploid invasive growth, and diploid pseudohyphal differentiation
Abstract
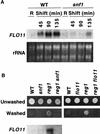
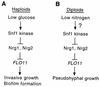
The Snf1 protein kinase of Saccharomyces cerevisiae is important for many cellular responses to glucose limitation, including haploid invasive growth. We show here that Snf1 regulates transcription of FLO11, which encodes a cell surface glycoprotein required for invasive growth. We further show that Nrg1 and Nrg2, two repressor proteins that interact with Snf1, function as negative regulators of invasive growth and as repressors of FLO11. We also examined the role of Snf1, Nrg1, and Nrg2 in two other Flo11-dependent processes. Mutations affected the initiation of biofilm formation, which is glucose sensitive, but also affected diploid pseudohyphal differentiation, thereby unexpectedly implicating Snf1 in a response to nitrogen limitation. Deletion of the NRG1 and NRG2 genes suppressed the defects of a snf1 mutant in all of these processes. These findings suggest a model in which the Snf1 kinase positively regulates Flo11-dependent developmental events by antagonizing Nrg-mediated repression of the FLO11 gene.
Figures



Similar articles
-
Role of the yeast Snf1 protein kinase in invasive growth.Biochem Soc Trans. 2003 Feb;31(Pt 1):175-7. doi: 10.1042/bst0310175. Biochem Soc Trans. 2003. PMID: 12546679 Review.
-
Snf1 kinases with different beta-subunit isoforms play distinct roles in regulating haploid invasive growth.Mol Cell Biol. 2003 Feb;23(4):1341-8. doi: 10.1128/MCB.23.4.1341-1348.2003. Mol Cell Biol. 2003. PMID: 12556493 Free PMC article.
-
Interaction of the repressors Nrg1 and Nrg2 with the Snf1 protein kinase in Saccharomyces cerevisiae.Genetics. 2001 Jun;158(2):563-72. doi: 10.1093/genetics/158.2.563. Genetics. 2001. PMID: 11404322 Free PMC article.
-
Repressors Nrg1 and Nrg2 regulate a set of stress-responsive genes in Saccharomyces cerevisiae.Eukaryot Cell. 2005 Nov;4(11):1882-91. doi: 10.1128/EC.4.11.1882-1891.2005. Eukaryot Cell. 2005. PMID: 16278455 Free PMC article.
-
Glucose repression in yeast.Curr Opin Microbiol. 1999 Apr;2(2):202-7. doi: 10.1016/S1369-5274(99)80035-6. Curr Opin Microbiol. 1999. PMID: 10322167 Review.
Cited by
-
Integrated analysis of transcriptome and lipid profiling reveals the co-influences of inositol-choline and Snf1 in controlling lipid biosynthesis in yeast.Mol Genet Genomics. 2012 Jul;287(7):541-54. doi: 10.1007/s00438-012-0697-5. Epub 2012 May 24. Mol Genet Genomics. 2012. PMID: 22622761
-
Deconstructing the genetic basis of spent sulphite liquor tolerance using deep sequencing of genome-shuffled yeast.Biotechnol Biofuels. 2015 Mar 31;8:53. doi: 10.1186/s13068-015-0241-z. eCollection 2015. Biotechnol Biofuels. 2015. PMID: 25866561 Free PMC article.
-
New Aspects of Invasive Growth Regulation Identified by Functional Profiling of MAPK Pathway Targets in Saccharomyces cerevisiae.Genetics. 2020 Sep;216(1):95-116. doi: 10.1534/genetics.120.303369. Epub 2020 Jul 14. Genetics. 2020. PMID: 32665277 Free PMC article.
-
Glucose signaling in Saccharomyces cerevisiae.Microbiol Mol Biol Rev. 2006 Mar;70(1):253-82. doi: 10.1128/MMBR.70.1.253-282.2006. Microbiol Mol Biol Rev. 2006. PMID: 16524925 Free PMC article. Review.
-
Roles of the Snf1-activating kinases during nitrogen limitation and pseudohyphal differentiation in Saccharomyces cerevisiae.Eukaryot Cell. 2010 Jan;9(1):208-14. doi: 10.1128/EC.00216-09. Epub 2009 Oct 30. Eukaryot Cell. 2010. PMID: 19880754 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
