The molybdate-responsive Escherichia coli ModE transcriptional regulator coordinates periplasmic nitrate reductase (napFDAGHBC) operon expression with nitrate and molybdate availability
- PMID: 12029041
- PMCID: PMC135098
- DOI: 10.1128/JB.184.12.3253-3259.2002
The molybdate-responsive Escherichia coli ModE transcriptional regulator coordinates periplasmic nitrate reductase (napFDAGHBC) operon expression with nitrate and molybdate availability
Abstract
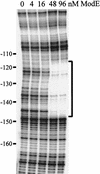
Expression of the Escherichia coli napFDAGHBC operon (also known as aeg46.5), which encodes the periplasmic molybdoenzyme for nitrate reduction, is increased in response to anaerobiosis and further stimulated by the addition of nitrate or to a lesser extent by nitrite to the cell culture medium. These changes are mediated by the transcription factors Fnr and NarP, respectively. Utilizing a napF-lacZ operon fusion, we demonstrate that napF gene expression is impaired in strain defective for the molybdate-responsive ModE transcription factor. This control abrogates nitrate- or nitrite-dependent induction during anaerobiosis. Gel shift and DNase I footprinting analyses establish that ModE binds to the napF promoter with an apparent K(d) of about 35 nM at a position centered at -133.5 relative to the start of napF transcription. Although the ModE binding site sequence is similar to other E. coli ModE binding sites, the location is atypical, because it is not centered near the start of transcription. Introduction of point mutations in the ModE recognition site severely reduced or abolished ModE binding in vitro and conferred a modE phenotype (i.e., loss of molybdate-responsive gene expression) in vivo. In contrast, deletion of the upstream ModE region site rendered napF expression independent of modE. These findings indicate the involvement of an additional transcription factor to help coordinate nitrate- and molybdate-dependent napF expression by the Fnr, NarP, NarL, and ModE proteins. The upstream ModE regulatory site functions to override nitrate control of napF gene expression when the essential enzyme component, molybdate, is limiting in the cell environment.
Figures




References
-
- Anderson, L. A., T. Palmer, N. C. Price, S. Borneman, D. H. Boxer, and R. N. Pau. 1997. Characterization of the molybdenum-responsive ModE regulatory protein and its binding to the promoter region of the modABCD (molybdenum transport) operon of Escherichia coli. Eur. J. Biochem. 246:119-126. - PubMed
-
- Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997.The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

