Microphthalmia-associated transcription factor interacts with LEF-1, a mediator of Wnt signaling
- PMID: 12032083
- PMCID: PMC126018
- DOI: 10.1093/emboj/21.11.2703
Microphthalmia-associated transcription factor interacts with LEF-1, a mediator of Wnt signaling
Abstract
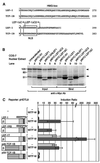
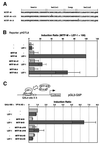
Wnt signals regulate differentiation of neural crest cells through the beta-catenin associated with a nuclear mediator of the lymphoid-enhancing factor 1 (LEF-1)/T-cell factors (TCFs) family. Here we show the interaction between the basic helix-loop-helix and leucine-zipper region of microphthalmia-associated transcription factor (MITF) and LEF-1. MITF is essential for melanocyte differentiation and its heterozygous mutations cause auditory-pigmentary syndromes. Functional cooperation of MITF with LEF-1 results in synergistic transactivation of the dopachrome tautomerase (DCT) gene promoter, an early melanoblast marker. This activation depends on the separate cis-acting elements, which are also responsible for the induction of the DCT promoter by lithium chloride that mimics Wnt signaling. beta-catenin is required for efficient transactivation, but dispensable for the interaction between MITF and LEF-1. The interaction with MITF is unique to LEF-1 and not detectable with TCF-1. LEF-1 also cooperates with the MITF-related proteins, such as TFE3, to transactivate the DCT promoter. This study therefore suggests that the MITF/TFE3 family is a new class of nuclear modulators for LEF-1, which may ensure efficient propagation of Wnt signals in many types of cells.
Figures







References
-
- Amae S. et al. (1998) Identification of a novel isoform of microphthalmia-associated transcription factor that is enriched in retinal pigment epithelium. Biochem. Biophys. Res. Commun., 247, 710–715. - PubMed
-
- Amae S., Yasumoto,K., Takeda,K., Udono,T., Takahashi,K. and Shibahara,S. (2000) Identification of a composite enhancer of the human tyrosinase-related protein 2/DOPAchrome tautomerase gene. Biochim. Biophys. Acta, 1492, 505–508. - PubMed
-
- Amiel J., Watkin,P.M., Tassabehji,M., Read,A.P. and Winter,R.M. (1998) Mutation of the MITF gene in albinism–deafness syndrome (Tietz syndrome). Clin. Dysmorphol., 7, 17–20. - PubMed
-
- Barker N., Morin,P.J. and Clevers,H. (2000) The yin-yang of TCF/ β-catenin signaling. Adv. Cancer Res., 77, 1–24. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

