Placental overgrowth in mice lacking the imprinted gene Ipl
- PMID: 12032310
- PMCID: PMC124258
- DOI: 10.1073/pnas.122039999
Placental overgrowth in mice lacking the imprinted gene Ipl
Abstract
The Ipl (Tssc3) gene lies in an extended imprinted region of distal mouse chromosome 7, which also contains the Igf2 gene. Expression of Ipl is highest in placenta and yolk sac, where its mRNA is derived almost entirely from the maternal allele. Ipl encodes a small cytoplasmic protein with a pleckstrin-homology (PH) domain. We constructed two lines of mice with germ-line deletions of this gene (Ipl(neo) and Ipl(loxP)) and another line deleted for the similar but nonimprinted gene Tih1. All three lines were viable. There was consistent overgrowth of the Ipl-null placentas, with expansion of the spongiotrophoblast. These larger placentas did not confer a fetal growth advantage; fetal size was normal in Ipl nulls with the Ipl(neo) allele and was decreased slightly in nulls with the Ipl(loxP) allele. When bred into an Igf2 mutant background, the Ipl deletion partially rescued the placental but not fetal growth deficiency. Neither fetal nor placental growth was affected by deletion of Tih1. These results show a nonredundant function for Ipl in restraining placental growth. The data further indicate that Ipl can act, at least in part, independently of insulin-like growth factor-2 signaling. Thus, genomic imprinting regulates multiple pathways to control placental size.
Figures



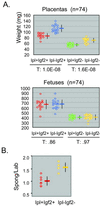
References
-
- Qian N, Frank D, O'Keefe D, Dao D, Zhao L, Yuan L, Wang Q, Keating M, Walsh C, Tycko B. Hum Mol Genet. 1997;6:2021–2029. - PubMed
-
- Lee M P, Feinberg A P. Cancer Res. 1998;58:1052–1056. - PubMed
-
- DeChiara T M, Robertson E J, Efstratiadis A. Cell. 1991;64:849–859. - PubMed
-
- Baker J, Liu J P, Robertson E J, Efstratiadis A. Cell. 1993;75:73–82. - PubMed
-
- Leighton P A, Ingram R S, Eggenschwiler J, Efstratiadis A, Tilghman S M. Nature (London) 1995;375:34–39. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

