Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments
- PMID: 12032340
- PMCID: PMC124316
- DOI: 10.1073/pnas.072210299
Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments
Abstract
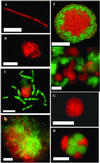
No microorganism capable of anaerobic growth on methane as the sole carbon source has yet been cultivated. Consequently, information about these microbes has been inferred from geochemical and microbiological observations of field samples. Stable isotope analysis of lipid biomarkers and rRNA gene surveys have implicated specific microbes in the anaerobic oxidation of methane (AOM). Here we use combined fluorescent in situ hybridization and secondary ion mass spectrometry analyses, to identify anaerobic methanotrophs in marine methane-seep sediments. The results provide direct evidence for the involvement of at least two distinct archaeal groups (ANME-1 and ANME-2) in AOM at methane seeps. Although both archaeal groups often occurred in direct physical association with bacteria, they also were observed as monospecific aggregations and as single cells. The ANME-1 archaeal group more frequently existed in monospecific aggregations or as single filaments, apparently without a bacterial partner. Bacteria associated with both archaeal groups included, but were not limited to, close relatives of Desulfosarcina species. Isotopic analyses suggest that monospecific archaeal cells and cell aggregates were active in anaerobic methanotrophy, as were multispecies consortia. In total, the data indicate that the microbial species and biotic interactions mediating anaerobic methanotrophy are diverse and complex. The data also clearly show that highly structured ANME-2/Desulfosarcina consortia are not the sole entities responsible for AOM at marine methane seeps. Other microbial groups, including ANME-1 archaea, are capable of anaerobic methane consumption either as single cells, in monospecific aggregates, or in multispecies consortia.
Figures


References
-
- Valentine D L, Reeburgh W S. Environ Microbiol. 2000;2:477–484. - PubMed
-
- Hinrichs K-U, Boetius A. In: Ocean Margin Systems. Wefer G, Billett D, Hebbeln D, Jørgensen B B, Schlüter M, van Weering T, editors. Heidelberg: Springer; 2002. , in press.
-
- Hinrichs K-U, Hayes J M, Sylva S P, Brewer P G, DeLong E F. Nature (London) 1999;398:802–805. - PubMed
-
- Hinrichs K-U, Summons R E, Orphan V J, Sylva S P, Hayes J M. Org Geochem. 2000;31:1685–1701.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

