An Arabidopsis gene encoding an alpha-xylosyltransferase involved in xyloglucan biosynthesis
- PMID: 12032363
- PMCID: PMC124356
- DOI: 10.1073/pnas.102644799
An Arabidopsis gene encoding an alpha-xylosyltransferase involved in xyloglucan biosynthesis
Abstract
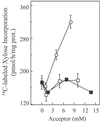
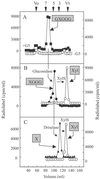
Microsomal membranes catalyze the formation of xyloglucan from UDP-Glc and UDP-Xyl by cooperative action of alpha-xylosyltransferase and beta-glucan synthase activities. Here we report that etiolated pea microsomes contain an alpha-xylosyltransferase that catalyzes the transfer of xylose from UDP-[(14)C]xylose onto beta(1,4)-linked glucan chains. The solubilized enzyme had the capacity to transfer xylosyl residues onto cello-oligosaccharides having 5 or more glucose residues. Analysis of the data from these biochemical assays led to the identification of a group of Arabidopsis genes and the hypothesis that one or more members of this group may encode alpha-xylosyltransferases involved in xyloglucan biosynthesis. To evaluate this hypothesis, the candidate genes were expressed in Pichia pastoris and their activities measured with the biochemical assay described above. One of the candidate genes showed cello-oligosaccharide-dependent xylosyltransferase activity. Characterization of the radiolabeled products obtained with cellopentaose as acceptor indicated that the pea and the Arabidopsis enzymes transfer the (14)C-labeled xylose mainly to the second glucose residue from the nonreducing end. Enzymatic digestion of these radiolabeled products produced results that would be expected if the xylose was attached in an alpha(1,6)-linkage to the glucan chain. We conclude that this Arabidopsis gene encodes an alpha-xylosyltransferase activity involved in xyloglucan biosynthesis.
Figures



References
-
- McCann M C, Roberts K. In: The Cytoskeletal Basis of Plant Growth and Form. Lloyd C W, editor. New York: Academic; 1991. pp. 109–120.
-
- Carpita N C, Gibeaut D M. Plant J. 1993;3:1–30. - PubMed
-
- Perrin R M, DeRocher A E, Bar-Peled M, Zeng W, Norambuena L, Orellana A, Raikhel N V, Keegstra K. Science. 1999;284:1976–1979. - PubMed
-
- Edwards M E, Dickson C A, Chengappa S, Sidebottom C, Gidley M J, Reid G. Plant J. 1999;19:691–697. - PubMed
-
- Faik A, Bar-Peled M, DeRocher A E, Zeng W, Perrin R M, Wilkerson C, Raikhel N V, Keegstra K. J Biol Chem. 2000;275:15082–15089. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

