The Rho-associated protein kinase p160ROCK is required for centrosome positioning
- PMID: 12034773
- PMCID: PMC2173415
- DOI: 10.1083/jcb.200203034
The Rho-associated protein kinase p160ROCK is required for centrosome positioning
Abstract
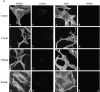
The p160-Rho-associated coiled-coil-containing protein kinase (ROCK) is identified as a new centrosomal component. Using immunofluorescence with a variety of p160ROCK antibodies, immuno EM, and depletion with RNA interference, p160ROCK is principally bound to the mother centriole (MC) and an intercentriolar linker. Inhibition of p160ROCK provoked centrosome splitting in G1 with the MC, which is normally positioned at the cell center and shows little motion during G1, displaying wide excursions around the cell periphery, similar to its migration toward the midbody during cytokinesis. p160ROCK inhibition late after anaphase in mitosis triggered MC migration to the midbody followed by completion of cell division. Thus, p160ROCK is required for centrosome positioning and centrosome-dependent exit from mitosis.
Figures









References
-
- Amano, M., Y. Fukata, and K. Kaibuchi. 2000. Regulation and functions of Rho-associated kinase. Exp. Cell Res. 261:44–51. - PubMed
-
- Andersen, S.S.L. 1999. Molecular characteristics of the centrosome. Int. Rev. Cytol. 187:51–109. - PubMed
-
- Bornens, M., M. Paintrand, J. Berges, M.C. Marty, and E. Karsenti. 1987. Structural and chemical characterization of isolated centrosomes. Cell Motil. Cytoskeleton. 8:238–249. - PubMed
-
- Chang, P., and T. Stearns. 2000. δ-Tubulin and ɛ-tubulin: two new human centrosomal tubulins reveal new aspects of centrosome structure and function. Nat. Cell Biol. 2:30–35. - PubMed
-
- Chevrier, V., S. Komesli, A.C. Schmit, M. Vantard, A.M. Lambert, and D. Job. 1992. A monoclonal antibody, raised against mammalian centrosomes and screened by recognition of plant microtubule organizing centers, identifies a pericentriolar component in different cell types. J. Cell Sci. 101:823–835. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

