A Tetrahymena thermophila ribozyme-based indicator gene to detect transposition of marked retroelements in mammalian cells
- PMID: 12034850
- PMCID: PMC117211
- DOI: 10.1093/nar/30.11.e49
A Tetrahymena thermophila ribozyme-based indicator gene to detect transposition of marked retroelements in mammalian cells
Abstract
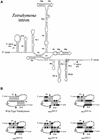
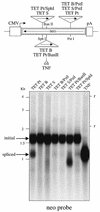
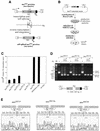
We devised an indicator gene for retrotransposition based on an autocatalytic ribozyme element--the Tetrahymena thermophila 23S rRNA group I intron--which can self-splice in vitro and does not require--at variance with nuclear mRNA introns--any specific pathway and cellular component for the completion of the splicing process. Several constructs, with the Tetrahymena intron adequately modified so as to be inserted at various positions within a neomycin-containing cassette under conditions that restore the neomycin-coding sequence after splicing out of the intron, were assayed for splicing efficiency in mammalian cells in culture. We show, both by northern blot analysis and by the recovery of neomycin activity upon retroviral transduction of the cassettes, that splicing efficiency depends on both the local base pairing and the global position of the intron within the neomycin transcript, and that some constructs are functional. We further show that they allow the efficient sorting out of retrotransposition events when assayed, as a control, with a human LINE retrotransposon. These indicator genes should be of great help in elucidating the mechanisms of transposition of a series of retroelements associated with transcripts not prone to nuclear mRNA intron splicing and previously not opened to any retrotransposition assay.
Figures


References
-
- Boeke J.D. and Stoye,J.P. (1997) Retrotransposons, endogenous retroviruses, and the evolution of retroelements. In Coffin,J.M., Hughes,S.H. and Varmus,H.E. (eds), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 343–435. - PubMed
-
- Heidmann O. and Heidmann,T. (1991) Retrotransposition of a mouse IAP sequence tagged with an indicator gene. Cell, 64, 159–170. - PubMed
-
- Moran J.V., Holmes,S.E., Nass,T.P., DeBerardinis,R.J., Boeke,J.D. and Kazazian,H.H.J. (1996) High frequency retroposition in cultured mammalian cells. Cell, 87, 917–927. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

