Mammalian mitogenomic relationships and the root of the eutherian tree
- PMID: 12034869
- PMCID: PMC123036
- DOI: 10.1073/pnas.102164299
Mammalian mitogenomic relationships and the root of the eutherian tree
Abstract
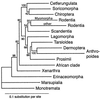
The strict orthology of mitochondrial (mt) coding sequences has promoted their use in phylogenetic analyses at different levels. Here we present the results of a mitogenomic study (i.e., analysis based on the set of protein-coding genes from complete mt genomes) of 60 mammalian species. This number includes 11 new mt genomes. The sampling comprises all but one of the traditional eutherian orders. The previously unrepresented order Dermoptera (flying lemurs) fell within Primates as the sister group of Anthropoidea, making Primates paraphyletic. This relationship was strongly supported. Lipotyphla ("insectivores") split into three distinct lineages: Erinaceomorpha, Tenrecomorpha, and Soricomorpha. Erinaceomorpha was the basal eutherian lineage. Sirenia (dugong) and Macroscelidea (elephant shrew) fell within the African clade. Pholidota (pangolin) joined the Cetferungulata as the sister group of Carnivora. The analyses identified monophyletic Pinnipedia with Otariidae (sea lions, fur seals) and Odobenidae (walruses) as sister groups to the exclusion of Phocidae (true seals).
Figures

References
-
- Arnason U, Johnsson E. J Mol Evol. 1992;34:493–505. - PubMed
-
- Mouchaty S K, Gullberg A, Janke A, Arnason U. Mol Biol Evol. 2000;17:60–67. - PubMed
-
- Mouchaty S K, Gullberg A, Janke A, Arnason U. Zool Scr. 2000;29:307–317.
-
- Madsen O, Scally M, Douady C J, Kao D J, DeBry R W, Adkins R, Amrine H M, Stanhope M J, de Jong W W, Springer M S. Nature (London) 2001;409:610–614. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

