Arabidopsis RAR1 exerts rate-limiting control of R gene-mediated defenses against multiple pathogens
- PMID: 12034891
- PMCID: PMC150601
- DOI: 10.1105/tpc.001040
Arabidopsis RAR1 exerts rate-limiting control of R gene-mediated defenses against multiple pathogens
Abstract
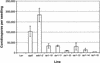
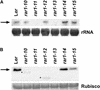
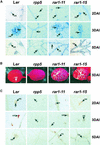
We have identified the Arabidopsis ortholog of barley RAR1 as a component of resistance specified by multiple nucleotide binding/Leu-rich repeat resistance (R) genes recognizing different bacterial and oomycete pathogen isolates. Characterization of partially and fully defective rar1 mutations revealed that wild-type RAR1 acts as a rate-limiting regulator of early R gene-triggered defenses, determining the extent of pathogen containment, hypersensitive plant cell death, and an oxidative burst at primary infection sites. We conclude that RAR1 defense signaling function is conserved between plant species that are separated evolutionarily by 150 million years. RAR1 encodes a protein with two zinc binding (CHORD) domains that are highly conserved across eukaryotic phyla, and the single nematode CHORD-containing homolog, Chp, was found previously to be essential for embryo viability. An absence of obvious developmental defects in null Arabidopsis rar1 mutants favors the notion that, in contrast, RAR1 does not play a fundamental role in plant development.
Figures


References
-
- Austin, M., Muskett, P.R., Kahn, K., Feys, B.J., Jones, J.D.G., and Parker, J.E. (2002). Regulatory role of SGT1 in early R gene-mediated plant defences. Science 295, 2077–2080. - PubMed
-
- Azevedo, C., Sadanandom, A., Kitagawa, K., Freialdenhoven, A., Shirasu, K., and Schulze-Lefert, P. (2002). The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295, 2073–2076. - PubMed
-
- Bent, A.F., Kunkel, B.N., Dahlbeck, D., Brown, K.L., Schmidt, R., Giraudat, J., Leung, J., and Staskawicz, B.J. (1994). RPS2 of Arabidopsis thaliana: A leucine-rich repeat class of plant disease resistance protein. Science 265, 1856–1860. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

