Development of a sensitive and specific enzyme-linked immunosorbent assay for detecting and quantifying CMY-2 and SHV beta-lactamases
- PMID: 12037047
- PMCID: PMC130713
- DOI: 10.1128/JCM.40.6.1947-1957.2002
Development of a sensitive and specific enzyme-linked immunosorbent assay for detecting and quantifying CMY-2 and SHV beta-lactamases
Abstract
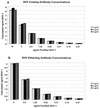
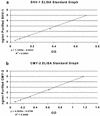
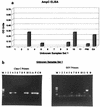
Polyclonal rabbit antibodies against SHV-1 and CMY-2 beta-lactamases were produced and characterized, and enzyme-linked immunosorbent assays (ELISAs) were developed. Immunoblots revealed that the anti-SHV-1 antibody recognized SHV-1 but did not recognize TEM-1, K-1, OXA-1, or any AmpC beta-lactamase tested. The anti-CMY-2 antibody detected Escherichia coli CMY-2, Enterobacter cloacae P99, Klebsiella pneumoniae ACT-1, and the AmpC beta-lactamases of Enterobacter aerogenes, Morganella morganii, and Citrobacter freundii. No cross-reactivity of the anti-CMY-2 antibody was seen against laboratory strains of E. coli possessing TEM-1, SHV-1, K-1, or OXA-1 beta-lactamases. Operating conditions for performing ELISAs were optimized. Both anti-CMY-2 and anti-SHV-1 antibodies detected picogram quantities of purified protein in ELISAs. The reactivity of the anti-CMY-2 antibody was tested against a number of AmpC beta-lactamases by assaying known quantities of purified enzymes in ELISAs (AmpC beta-lactamases of M. morganii, C. freundii, E. coli, and E. cloacae). As the homology to CMY-2 beta-lactamase decreased, the minimum level needed for detection increased (e.g., 94% homology recognized at 1 ng/ml and 71% homology recognized at 10 ng/ml). The ELISAs were used to assay unknown clinical isolates for AmpC and SHV beta-lactamases, and the results were confirmed with PCR amplification of bla(AmpC) and bla(SHV) genes. Overall, we found that our ELISAs were at least 95% sensitive and specific for detecting SHV and AmpC beta-lactamases. The ELISA format can facilitate the identification of AmpC and SHV beta-lactamases and can be used to quantify relative amounts of beta-lactamase enzymes in clinical and laboratory isolates.
Figures

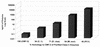
References
-
- Barker, L. R., and J. C. Roberts. 1994. The diagnostic process, p. 8-10. In L. R. Barker, J. R. Burton, and P. D. Zieve (ed.), Principles of ambulatory medicine, 4th ed. The Williams & Wilkins Co., Baltimore, Md.
-
- Bauernfeind, A., Y. Chong, and K. Lee. 1998. Plasmid-encoded AmpC β-lactamases: how far have we gone 10 years after the discovery? Yonsei Med. J. 39:520-525. - PubMed
-
- Bibi, E., and R. Laskov. 1990. Selection and application of antibodies modifying the function of β-lactamase. Biochim. Biophys. Acta 1035:237-241. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

