Automated ribotyping of vancomycin-resistant Enterococcus faecium isolates
- PMID: 12037051
- PMCID: PMC130812
- DOI: 10.1128/JCM.40.6.1977-1984.2002
Automated ribotyping of vancomycin-resistant Enterococcus faecium isolates
Abstract
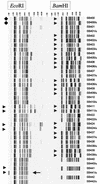
Vancomycin-resistant Enterococcus faecium (VREF) strains represent an important threat in hospital infections in the United States and are found at high frequencies in both the community and farm animals in Europe. We evaluated automated ribotyping for interlaboratory reproducibility by using the restriction enzymes EcoRI and BamHI and compared ribotyping to both amplification of fragment length polymorphism (AFLP) analysis and multilocus sequence typing (MLST) to assess its discriminatory power and capacity for the identification of epidemiologically important strains. Of 19 (EcoRI) and 16 (BamHI) isolates tested in duplicate in two laboratories, 18 (95%) and 16 (100%), respectively, showed reproducible ribotypes. These high reproducibility rates were obtained only after manual refinement of the automated fingerprint analysis. A group of 49 VREF strains initially selected to represent 32 distinct AFLP types were separated into 28 EcoRI ribotypes, 25 BamHI ribotypes, and 28 sequence types. Ribotyping with EcoRI and BamHI was able to discern the host-specific genogroups recently disclosed by AFLP typing and MLST and to distinguish most strains containing the esp gene, a marker specific for strains causing hospital outbreaks. An expandable ribotype identification library was created. We recommend EcoRI as the enzyme of choice for automated ribotyping of VREF strains. Given the high level of discrimination of VREF strains, the high rate of interlaboratory reproducibility, and the potential for the identification of epidemiologically important genotypes, automated ribotyping appears to be a very valuable approach for characterizing VREF strains.
Figures

References
-
- Antonishyn, N. A., R. R. McDonald, E. L. Chan, G. Horsman, C. E. Woodmansee, P. S. Falk, and C. G. Mayhall. 2000. Evaluation of fluorescence-based amplified fragment length polymorphism analysis for molecular typing in hospital epidemiology: comparison with pulsed-field gel electrophoresis for typing strains of vancomycin-resistant Enterococcus faecium. J. Clin. Microbiol. 38:4058-4065. - PMC - PubMed
-
- Brisse, S., A. C. Fluit, U. Wagner, P. Heisig, D. Milatovic, J. Verhoef, S. Scheuring, K. Kohrer, and F. J. Schmitz. 1999. Association of alterations in ParC and GyrA proteins with resistance of clinical isolates of Enterococcus faecium to nine different fluoroquinolones. Antimicrob. Agents Chemother. 43:2513-2516. - PMC - PubMed
-
- Bruce, J. L. 1996. Automated system rapidly identifies and characterizes microorganisms in food. Food Technol. 50:77-81.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

