oriC region and replication termination site, dif, of the Xanthomonas campestris pv. campestris 17 chromosome
- PMID: 12039751
- PMCID: PMC123971
- DOI: 10.1128/AEM.68.6.2924-2933.2002
oriC region and replication termination site, dif, of the Xanthomonas campestris pv. campestris 17 chromosome
Abstract
A 13-kb DNA fragment containing oriC and the flanking genes thdF, orf900, yidC, rnpA, rpmH, oriC, dnaA, dnaN, recF, and gyrB was cloned from the gram-negative plant pathogen Xanthomonas campestris pv. campestris 17. These genes are conserved in order with other eubacterial oriC genes and code for proteins that share high degrees of identity with their homologues, except for orf900, which has a homologue only in Xylella fastidiosa. The dnaA/dnaN intergenic region (273 bp) identified to be the minimal oriC region responsible for autonomous replication has 10 pure AT clusters of four to seven bases and only three consensus DnaA boxes. These findings are in disagreement with the notion that typical oriCs contain four or more DnaA boxes located upstream of the dnaA gene. The X. campestris pv. campestris 17 attB site required for site-specific integration of cloned fragments from filamentous phage phiLf replicative form DNA was identified to be a dif site on the basis of similarities in nucleotide sequence and function with the Escherichia coli dif site required for chromosome dimer resolution and whose deletion causes filamentation of the cells. The oriC and dif sites were located at 12:00 and 6:00, respectively, on the circular X. campestris pv. campestris 17 chromosome map, similar to the locations found for E. coli sites. Computer searches revealed the presence of both the dif site and XerC/XerD recombinase homologues in 16 of the 42 fully sequenced eubacterial genomes, but eight of the dif sites are located far away from the 6:00 point instead of being placed opposite the cognate oriC. The differences in the relative position suggest that mechanisms different from that of E. coli may participate in the control of chromosome replication.
Figures


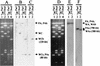



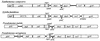
References
-
- Blakely, G., S. Colloms, G. May, M. Burke, and D. Sherratt. 1991. Escherichia coli XerC recombinase is required for chromosomal segregation at cell division. New Biol. 3:789-798. - PubMed
-
- Blakely, G., G. May, R. McCulloch, L. K. Arciszewska, M. Burke, S. T. Lovett, and D. J. Sherratt. 1993. Two related recombinases are required for site-specific recombination at dif and cer in E. coli K12. Cell 75:351-361. - PubMed
-
- Bradbury, J. F. 1984. Genus II. Xanthomonas Dowson 1939:187.AL, p. 199. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology. Williams & Wilkins, Baltimore, Md.
-
- Clerget, M. 1991. Site-specific recombination promoted by a short DNA segment of plasmid R1 and by a homologous segment in the terminus region of the Escherichia coli chromosome. New Biol. 3:780-788. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

