Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria
- PMID: 12039771
- PMCID: PMC123953
- DOI: 10.1128/AEM.68.6.3094-3101.2002
Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria
Abstract
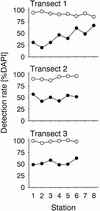
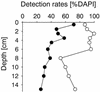
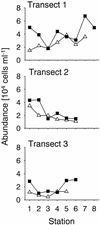
Fluorescence in situ hybridization (FISH) with horseradish peroxidase (HRP)-labeled oligonucleotide probes and tyramide signal amplification, also known as catalyzed reporter deposition (CARD), is currently not generally applicable to heterotrophic bacteria in marine samples. Penetration of the HRP molecule into bacterial cells requires permeabilization procedures that cause high and most probably species-selective cell loss. Here we present an improved protocol for CARD-FISH of marine planktonic and benthic microbial assemblages. After concentration of samples onto membrane filters and subsequent embedding of filters in low-gelling-point agarose, no decrease in bacterial cell numbers was observed during 90 min of lysozyme incubation (10 mg ml(-1) at 37 degrees C). The detection rates of coastal North Sea bacterioplankton by CARD-FISH with a general bacterial probe (EUB338-HRP) were significantly higher (mean, 94% of total cell counts; range, 85 to 100%) than that with a monolabeled probe (EUB338-mono; mean, 48%; range, 19 to 66%). Virtually no unspecific staining was observed after CARD-FISH with an antisense EUB338-HRP. Members of the marine SAR86 clade were undetectable by FISH with a monolabeled probe; however, a substantial population was visualized by CARD-FISH (mean, 7%; range, 3 to 13%). Detection rates of EUB338-HRP in Wadden Sea sediments (mean, 81%; range, 53 to 100%) were almost twice as high as the detection rates of EUB338-mono (mean, 44%; range, 25 to 71%). The enhanced fluorescence intensities and signal-to-background ratios make CARD-FISH superior to FISH with directly labeled oligonucleotides for the staining of bacteria with low rRNA content in the marine environment.
Figures



References
-
- Bobrow, M. N., T. D. Harris, K. J. Shaughnessy, and G. J. Litt. 1989. Catalyzed reporter deposition, a novel method of signal amplification: application to immunoassays. J. Immunol. Methods 125:279-285. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

