RNA editing by adenosine deaminases that act on RNA
- PMID: 12045112
- PMCID: PMC1823043
- DOI: 10.1146/annurev.biochem.71.110601.135501
RNA editing by adenosine deaminases that act on RNA
Abstract
ADARs are RNA editing enzymes that target double-stranded regions of nuclear-encoded RNA and viral RNA. These enzymes are particularly abundant in the nervous system, where they diversify the information encoded in the genome, for example, by altering codons in mRNAs. The functions of ADARs in known substrates suggest that the enzymes serve to fine-tune and optimize many biological pathways, in ways that we are only starting to imagine. ADARs are also interesting in regard to the remarkable double-stranded structures of their substrates and how enzyme specificity is achieved with little regard to sequence. This review summarizes ongoing investigations of the enzyme family and their substrates, focusing on biological function as well as biochemical mechanism.
Figures
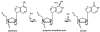




References
-
- Bass BL, editor. RNA Editing. Oxford: Oxford Univ. Press; 2001. p. 210.
-
- Gott JM, Emeson RB. Annu Rev Genet. 2000;34:499–531. - PubMed
-
- Benne R, Van den Burg J, Brakenhoff JP, Sloof P, Van Boom JH, et al. Cell. 1986;46:819–26. - PubMed
-
- Powell LM, Wallis SC, Pease RJ, Edwards YH, Knott TJ, et al. Cell. 1987;50:831–40. - PubMed
-
- Gerber AP, Keller W. Trends Biochem Sci. 2001;26:376–84. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

