Compositional gradients in Gramineae genes
- PMID: 12045139
- PMCID: PMC1383739
- DOI: 10.1101/gr.189102
Compositional gradients in Gramineae genes
Abstract
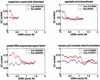
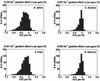
In this study, we describe a property of Gramineae genes, and perhaps all monocot genes, that is not observed in eudicot genes. Along the direction of transcription, beginning at the junction of the 5'-UTR and the coding region, there are gradients in GC content, codon usage, and amino-acid usage. The magnitudes of these gradients are large enough to hinder the annotation of the rice genome and to confound the detection of protein homologies across the monocot-eudicot divide.
Figures






References
-
- Altschul SF, Gish W. Local alignment statistics. Methods Enzymol. 1996;266:460–480. - PubMed
-
- Bernardi G. Isochores and the evolutionary genomics of vertebrates. Gene. 2000;241:3–17. - PubMed
-
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. - PubMed
-
- Chothia C. The nature of the accessible and buried surfaces in proteins. J Mol Biol. 1976;105:1–12. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
