Fourfold faster rate of genome rearrangement in nematodes than in Drosophila
- PMID: 12045140
- PMCID: PMC1383740
- DOI: 10.1101/gr.172702
Fourfold faster rate of genome rearrangement in nematodes than in Drosophila
Abstract
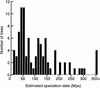
We compared the genome of the nematode Caenorhabditis elegans to 13% of that of Caenorhabditis briggsae, identifying 252 conserved segments along their chromosomes. We detected 517 chromosomal rearrangements, with the ratio of translocations to inversions to transpositions being approximately 1:1:2. We estimate that the species diverged 50-120 million years ago, and that since then there have been 4030 rearrangements between their whole genomes. Our estimate of the rearrangement rate, 0.4-1.0 chromosomal breakages/Mb per Myr, is at least four times that of Drosophila, which was previously reported to be the fastest rate among eukaryotes. The breakpoints of translocations are strongly associated with dispersed repeats and gene family members in the C. elegans genome.
Figures
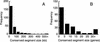




Comment in
-
Genomes in motion.Genome Res. 2002 Jun;12(6):843. doi: 10.1101/gr.293102. Genome Res. 2002. PMID: 12045137 No abstract available.
References
-
- Aguinaldo AM, Turbeville JM, Linford LS, Rivera MC, Garey JR, Raff RA, Lake JA. Evidence for a clade of nematodes, arthropods and other moulting animals. Nature. 1997;387:489–493. - PubMed
-
- Baillie DL, Rose AM. WABA success: A tool for sequence comparison between large genomes. Genome Res. 2000;10:1071–1073. - PubMed
-
- Baird S, Sutherlin ME, Emmons SW. Reproductive isolation in Rhabditidae (Nematoda: Secernentea); mechanisms that isolate six species of three genera. Evolution. 1992;46:585–594. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
