Characterization of Tbc2, a nucleus-encoded factor specifically required for translation of the chloroplast psbC mRNA in Chlamydomonas reinhardtii
- PMID: 12045185
- PMCID: PMC2174033
- DOI: 10.1083/jcb.200201060
Characterization of Tbc2, a nucleus-encoded factor specifically required for translation of the chloroplast psbC mRNA in Chlamydomonas reinhardtii
Abstract
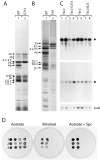
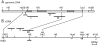
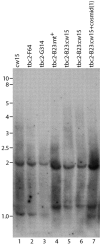
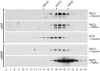
Genetic analysis has revealed that the three nucleus-encoded factors Tbc1, Tbc2, and Tbc3 are involved in the translation of the chloroplast psbC mRNA of the eukaryotic green alga Chlamydomonas reinhardtii. In this study we report the isolation and phenotypic characterization of two new tbc2 mutant alleles and their use for cloning and characterizing the Tbc2 gene by genomic complementation. TBC2 encodes a protein of 1,115 residues containing nine copies of a novel degenerate 38-40 amino acid repeat with a quasiconserved PPPEW motif near its COOH-terminal end. The middle part of the Tbc2 protein displays partial amino acid sequence identity with Crp1, a protein from Zea mays that is implicated in the processing and translation of the chloroplast petA and petD RNAs. The Tbc2 protein is enriched in chloroplast stromal subfractions and is associated with a 400-kD protein complex that appears to play a role in the translation of specifically the psbC mRNA.
Figures


References
-
- Barkan, A., and M. Goldschmidt-Clermont. 2000. Participation of nuclear genes in chloroplast gene expression. Biochimie. 82:559–572. - PubMed
-
- Bennoun, P., M. Spierer-Herz, J. Erickson, J. Girard-Bascou, Y. Pierre, M. Delsome, and J.-D. Rochaix. 1986. Characterization of photosystem II mutants of Chlamydomonas reinhardtii lacking the psbA gene. Plant Mol. Biol. 6:151–160. - PubMed
-
- Blatch, G.L., and M. Lassle. 1999. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays. 21:932–939. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

